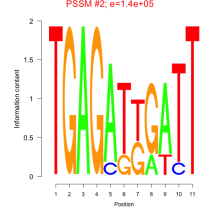
Thaps_bicluster_0275 Residual: 0.38
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0275 | 0.38 | Thalassiosira pseudonana |
Displaying 1 - 28 of 28
" class="views-fluidgrid-wrapper clear-block">
10013 hypothetical protein
10918 hypothetical protein
10945 hypothetical protein
19289 (Tp_CBF/NF3) CBFD_NFYB_HMF
21357 hypothetical protein
22071 Ion_trans
22979 hypothetical protein
23160 PLDc_Nuc_like_unchar1_1
23264 hypothetical protein
25435 hypothetical protein
25525 hypothetical protein
261002 WD40 superfamily
269151 hypothetical protein
269520 (CGL1) MetC
2711 hypothetical protein
2893 hypothetical protein
31406 (ARF4) Arl3
3554 hypothetical protein
37120 Yippee-Mis18
7776 hypothetical protein
8281 hypothetical protein
8308 hypothetical protein
8475 hypothetical protein
8611 VCBS
896 hypothetical protein
9008 hypothetical protein
9236 hypothetical protein
9375 (Tp_HSF_4d) HSF_DNA-bind
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006816 | calcium ion transport | 0.0135745 | 1 | Thaps_bicluster_0275 |
| GO:0006811 | ion transport | 0.0402221 | 1 | Thaps_bicluster_0275 |
| GO:0006520 | amino acid metabolism | 0.0467809 | 1 | Thaps_bicluster_0275 |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 0.0467809 | 1 | Thaps_bicluster_0275 |
| GO:0007264 | small GTPase mediated signal transduction | 0.0747343 | 1 | Thaps_bicluster_0275 |
| GO:0006812 | cation transport | 0.0936478 | 1 | Thaps_bicluster_0275 |
| GO:0006886 | intracellular protein transport | 0.116287 | 1 | Thaps_bicluster_0275 |
| GO:0000074 | regulation of cell cycle | 0.128417 | 1 | Thaps_bicluster_0275 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.151344 | 1 | Thaps_bicluster_0275 |
| GO:0008152 | metabolism | 0.620117 | 1 | Thaps_bicluster_0275 |


Comments