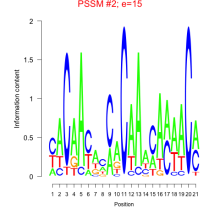
Thaps_bicluster_0280 Residual: 0.36
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0280 | 0.36 | Thalassiosira pseudonana |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
10340 hypothetical protein
11693 tRNA_SAD superfamily
16507 PLN02824
19435 PTZ00184
20622 PRK11728 superfamily
2107 hypothetical protein
21421 hypothetical protein
21603 hypothetical protein
21753 hypothetical protein
21771 hypothetical protein
22521 hypothetical protein
22850 DUF4239
2293 hypothetical protein
24197 hypothetical protein
25016 ALDH-SF superfamily
261228 Robl_LC7
30846 DUF2997
31232 (PFK1) PFK superfamily
3387 hypothetical protein
34511 PRK05862
4121 hypothetical protein
5291 hypothetical protein
7029 SET
7140 Ifi-6-16
7977 hypothetical protein
8347 hypothetical protein
8675 DUF4336 superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009613 | response to pest, pathogen or parasite | 0.00927255 | 1 | Thaps_bicluster_0280 |
| GO:0006096 | glycolysis | 0.0376815 | 1 | Thaps_bicluster_0280 |
| GO:0008152 | metabolism | 0.0595321 | 1 | Thaps_bicluster_0280 |
| GO:0006118 | electron transport | 0.24643 | 1 | Thaps_bicluster_0280 |


Comments