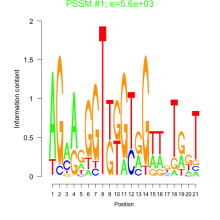
Thaps_bicluster_0281 Residual: 0.33
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0281 | 0.33 | Thalassiosira pseudonana |
Displaying 1 - 19 of 19
" class="views-fluidgrid-wrapper clear-block">
10050 DUF2233
10244 hypothetical protein
10818 tRNA_bindingDomain superfamily
10869 Sm_like superfamily
10886 hypothetical protein
11332 crotonase-like
11744 AdoMet_MTases superfamily
1465 TLDc
1649 COG0790
1911 hypothetical protein
2045 hypothetical protein
22877 SapB
5957 hypothetical protein
6234 DUF4239
6817 hypothetical protein
7602 hypothetical protein
8414 NA
8599 hypothetical protein
9834 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006412 | translation | 0.0200978 | 1 | Thaps_bicluster_0281 |
| GO:0006310 | DNA recombination | 0.0271911 | 1 | Thaps_bicluster_0281 |
| GO:0006397 | mRNA processing | 0.031426 | 1 | Thaps_bicluster_0281 |
| GO:0006281 | DNA repair | 0.0647398 | 1 | Thaps_bicluster_0281 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.0715551 | 1 | Thaps_bicluster_0281 |
| GO:0006468 | protein amino acid phosphorylation | 0.270534 | 1 | Thaps_bicluster_0281 |
| GO:0008152 | metabolism | 0.459719 | 1 | Thaps_bicluster_0281 |


Comments