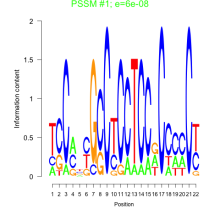
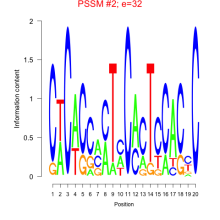
Thaps_bicluster_0286 Residual: 0.31
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0286 | 0.31 | Thalassiosira pseudonana |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
10041 hypothetical protein
16679 RRM_PPIL4
1693 hypothetical protein
19631 EFh
20578 hypothetical protein
22249 ATP12
22690 RRM_Nop6
23144 hypothetical protein
24598 Peptidase_C19
262617 (Tp_Myb2R7) MYB DNA binding protein/ transcription factor-like protein
264386 (Tp_Myb1R8) MYB DNA binding protein/ transcription factor-like protein
264498 PRK00142
268655 HDAC_classII_2
269723 hypothetical protein
3026 PseudoU_synth superfamily
3049 hypothetical protein
32713 DnaJ
33182 S_TKc
35929 (MAPKKK) S_TKc
4038 hypothetical protein
5266 hypothetical protein
6265 hypothetical protein
6591 GluZincin superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006457 | protein folding | 0.013778 | 1 | Thaps_bicluster_0286 |
| GO:0006468 | protein amino acid phosphorylation | 0.0350131 | 1 | Thaps_bicluster_0286 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.0715551 | 1 | Thaps_bicluster_0286 |
| GO:0006396 | RNA processing | 0.081025 | 1 | Thaps_bicluster_0286 |
| GO:0006508 | proteolysis and peptidolysis | 0.349482 | 1 | Thaps_bicluster_0286 |

Comments