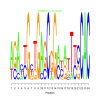
Module 42 Residual: 0.58
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0042 | v02 | 0.58 | -16.93 |
| Rv1069c Predicted membrane protein (DUF2319) | Rv1072 PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| Rv1557 Transmembrane transport protein MmpL6 | Rv1558 AclJ |
| Rv1781c 4-alpha-glucanotransferase (amylomaltase) (EC 2.4.1.25) | Rv1782 Possible membrane protein |
| Rv1783 Type VII secretion protein EccCa, FtsK/SpoIIIE family | Rv2486 Enoyl-CoA hydratase (EC 4.2.1.17) |
| Rv2968c PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN | Rv2969c POSSIBLE CONSERVED MEMBRANE OR SECRETED PROTEIN |
| Rv3837c putative phosphoglycerate mutase | Rv3838c Prephenate dehydratase (EC 4.2.1.51) |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0005887 | integral to plasma membrane |
|
GO:0005576 | extracellular region |
|
GO:0010033 | response to organic substance |
|
GO:0008150 | biological_process |
|
GO:0004300 | enoyl-CoA hydratase activity |
|
GO:0008201 | heparin binding |
|
GO:0040007 | growth |
|
GO:0051701 | interaction with host |
|
GO:0009986 | cell surface |
|
GO:0004664 | prephenate dehydratase activity |
|
GO:0016597 | amino acid binding |
|
GO:0033585 | L-phenylalanine biosynthetic process from chorismate via phenylpyruvate |
|
GO:0042803 | protein homodimerization activity |
|
GO:0051289 | protein homotetramerization |
|
GO:0004106 | chorismate mutase activity |















Discussion