Module 44 Residual: 0.54
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0044 | v02 | 0.54 | -12.43 |
| Rv1138c PUTATIVE OXIDOREDUCTASE | Rv1267c Transcriptional regulatory protein EmbR |
| Rv1521 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) | Rv1651c PE family protein |
| Rv2670c putative ATP/GTP-binding integral membrane protein | Rv3113 hydrolase, haloacid dehalogenase-like family |
| Rv3114 | Rv3126c |
| Rv3127 Oxygen-insensitive NADPH nitroreductase (EC 1.-.-.-) | Rv3130c Wax ester synthase/acyl-CoA:diacylglycerol acyltransferase |
| Rv3131 Dinucleotide-utilizing enzymes involved in molybdopterin and thiamine biosynthesis family 2 | Rv3168 Phosphotransferase |
| Rv3169 Putative uncharacterized protein BCG_3193 | Rv3253c Amino acid permease |
| Rv1090 PROBABLE CELLULASE CELA2B (ENDO-1,4-BETA-GLUCANASE) (ENDOGLUCANASE) (CARBOXYMETHYL CELLULASE) (EC 3.2.1.4) |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0003677 | DNA binding |
|
GO:0003924 | GTPase activity |
|
GO:0005886 | plasma membrane |
|
GO:0016887 | ATPase activity |
|
GO:0040007 | growth |
|
GO:0001666 | response to hypoxia |
|
GO:0004144 | diacylglycerol O-acyltransferase activity |
|
GO:0019432 | triglyceride biosynthetic process |
|
GO:0045017 | glycerolipid biosynthetic process |
|
GO:0047196 | long-chain-alcohol O-fatty-acyltransferase activity |
|
GO:0071731 | response to nitric oxide |
|
GO:0071500 | cellular response to nitrosative stress |

















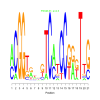
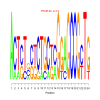
Discussion