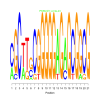
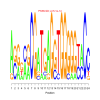
Module 61 Residual: 0.63
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0061 | v02 | 0.63 | -23.73 |
| Rv0226c Possible membrane protein | Rv0227c PROBABLE CONSERVED MEMBRANE PROTEIN |
| Rv0491 Phosphate regulon transcriptional regulatory protein PhoB (SphR); Sensory transduction protein regX3 | Rv1560 |
| Rv1561 | Rv1883c |
| Rv1884c Resuscitation-promoting factor RpfC | Rv1885c Periplasmic chorismate mutase I precursor (EC 5.4.99.5) |
| Rv2068c Beta-lactamase class A | Rv2069 RNA polymerase sigma-54 factor RpoN |
| Rv2413c DNA polymerase III delta subunit (EC 2.7.7.7) | Rv2696c |
| Rv3213c Chromosome (plasmid) partitioning protein ParA / Sporulation initiation inhibitor protein Soj | Rv3214 putative phosphoglycerate mutase family protein |
| Rv3215 Isochorismate synthase (EC 5.4.4.2) | Rv3693 Cell division protein DivIC (FtsB), stabilizes FtsL against RasP cleavage |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0016853 | isomerase activity |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0005887 | integral to plasma membrane |
|
GO:0008150 | biological_process |
|
GO:0003677 | DNA binding |
|
GO:0009405 | pathogenesis |
|
GO:0006355 | regulation of transcription, DNA-dependent |
|
GO:0015643 | toxin binding |
|
GO:0045727 | positive regulation of translation |
|
GO:0045927 | positive regulation of growth |
|
GO:0075136 | response to host |
|
GO:0004540 | ribonuclease activity |
|
GO:0017148 | negative regulation of translation |
|
GO:0045926 | negative regulation of growth |
|
GO:0005576 | extracellular region |
|
GO:0010628 | positive regulation of gene expression |
|
GO:0010629 | negative regulation of gene expression |
|
GO:0040010 | positive regulation of growth rate |
|
GO:0004106 | chorismate mutase activity |
|
GO:0005737 | cytoplasm |
|
GO:0042803 | protein homodimerization activity |
|
GO:0008800 | beta-lactamase activity |
|
GO:0030655 | beta-lactam antibiotic catabolic process |
|
GO:0033250 | penicillinase activity |
|
GO:0033251 | cephalosporinase activity |
|
GO:0046677 | response to antibiotic |
|
GO:0001666 | response to hypoxia |
|
GO:0009408 | response to heat |
|
GO:0009410 | response to xenobiotic stimulus |
|
GO:0009415 | response to water stimulus |
|
GO:0005829 | cytosol |
|
GO:0005618 | cell wall |
|
GO:0004619 | phosphoglycerate mutase activity |
|
GO:0003993 | acid phosphatase activity |
|
GO:0008909 | isochorismate synthase activity |


















Discussion