Module 143 Residual: 0.53
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0143 | v02 | 0.53 | -12.38 |
| Rv0081 transcriptional regulator, ArsR family | Rv0082 Formate hydrogenlyase subunit 11 |
| Rv0083 Hydrogenase-4 component B (EC 1.-.-.-) / Formate hydrogenlyase subunit 3 | Rv0084 Formate hydrogenlyase subunit 4 |
| Rv0085 Hydrogenase-4 component E (EC 1.-.-.-) | Rv0086 Hydrogenase-4 component F (EC 1.-.-.-) |
| Rv0776c | Rv0777 Adenylosuccinate lyase (EC 4.3.2.2) |
| Rv0778 Cytochrome P450 126A3 | Rv0845 POSSIBLE TWO COMPONENT SENSOR KINASE |
| Rv1159 probable conserved membrane protein | Rv1235 Maltose/maltodextrin ABC transporter, substrate binding periplasmic protein MalE |
| Rv1236 Maltose/maltodextrin ABC transporter, permease protein MalF | Rv1921c Lipoprotein LppF |
| Rv1922 D-alanyl-D-alanine carboxypeptidase (EC 3.4.16.4) | Rv1923 Lipase LipD |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0045333 | GO:0015980 | GO:0006091 | GO:0055114 | GO:0003954 | GO:0016655 | GO:0016651 | cellular respiration | energy derivation by oxidation of organi... | generation of precursor metabolites and ... | oxidation-reduction process | NADH dehydrogenase activity | oxidoreductase activity, acting on NAD(P... | oxidoreductase activity, acting on NAD(P... |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0004018 | N6-(1,2-dicarboxyethyl)AMP AMP-lyase (fumarate-forming) activity |
|
GO:0005618 | cell wall |
|
GO:0009247 | glycolipid biosynthetic process |
|
GO:0044117 | growth of symbiont in host |


















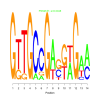
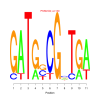
Discussion