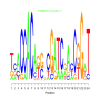
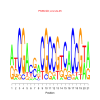
Module 218 Residual: 0.55
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0218 | v02 | 0.55 | -23.39 |
| Rv0069c L-serine dehydratase (EC 4.3.1.17) | Rv0070c Serine hydroxymethyltransferase (EC 2.1.2.1) |
| Rv0500 Pyrroline-5-carboxylate reductase (EC 1.5.1.2) | Rv1141c Enoyl-CoA hydratase (EC 4.2.1.17) |
| Rv1143 Alpha-methylacyl-CoA racemase (EC 5.1.99.4) | Rv1144 3-hydroxyacyl-CoA dehydrogenase [isoleucine degradation] (EC 1.1.1.35) |
| Rv1261c | Rv1262c Bis(5'-nucleosyl)-tetraphosphatase (asymmetrical) (EC 3.6.1.17) |
| Rv1556 Transcriptional regulator, TetR family | Rv3788 Nucleoside diphosphate kinase regulator |
| Rv0739 | Rv1142c Enoyl-CoA hydratase (EC 4.2.1.17) |
| Rv1896c Putative S-adenosyl-L-methionine-dependent methyltransferase | Rv3787c |
|
GO:0005886 | plasma membrane |
|
GO:0004372 | glycine hydroxymethyltransferase activity |
|
GO:0004735 | pyrroline-5-carboxylate reductase activity |
|
GO:0040007 | growth |
|
GO:0004300 | enoyl-CoA hydratase activity |
|
GO:0008111 | alpha-methylacyl-CoA racemase activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0005829 | cytosol |
















Discussion