Module 251 Residual: 0.46
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0251 | v02 | 0.46 | -9.31 |
| Rv0443 | Rv0558 Ubiquinone/menaquinone biosynthesis methyltransferase UbiE (EC 2.1.1.-) @ 2-heptaprenyl-1,4-naphthoquinone methyltransferase (EC 2.1.1.163) |
| Rv1229c Mrp protein homolog | Rv3376 Hydrolase, haloacid dehalogenase-like family |
| Rv2334 Cysteine synthase (EC 2.5.1.47) | Rv2335 Serine acetyltransferase (EC 2.3.1.30) |
| Rv3396c GMP synthase [glutamine-hydrolyzing] (EC 6.3.5.2) | Rv3684 Cysteine synthase (EC 2.5.1.47) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006534 | GO:0006563 | GO:0009070 | GO:0000097 | GO:0044283 | GO:0044711 | GO:0000096 | GO:1901607 | cysteine metabolic process | L-serine metabolic process | serine family amino acid biosynthetic pr... | sulfur amino acid biosynthetic process | small molecule biosynthetic process | single-organism biosynthetic process | sulfur amino acid metabolic process | alpha-amino acid biosynthetic process |
|
GO:0008168 | methyltransferase activity |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0004124 | cysteine synthase activity |
|
GO:0005515 | protein binding |
|
GO:0005576 | extracellular region |
|
GO:0030170 | pyridoxal phosphate binding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0009001 | serine O-acetyltransferase activity |
|
GO:0003922 | GMP synthase (glutamine-hydrolyzing) activity |
|
GO:0005829 | cytosol |










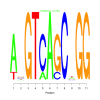

Discussion