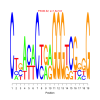
Module 269 Residual: 0.48
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0269 | v02 | 0.48 | -16.57 |
| Rv0168 | Rv0177 FIG034772: Probable conserved MCE associated protein |
| Rv0178 FIG030769: Probable conserved MCE associated membrane protein | Rv0249c putative succinate dehydrogenase [membrane anchor subunit] (succinic dehydrogenase) |
| Rv0933 Phosphate transport ATP-binding protein PstB (TC 3.A.1.7.1) | Rv0934 Phosphate ABC transporter, periplasmic phosphate-binding protein PstS (TC 3.A.1.7.1) |
| Rv0935 Phosphate transport system permease protein PstC (TC 3.A.1.7.1) | Rv0936 Phosphate transport system permease protein PstA (TC 3.A.1.7.1) |
| Rv1612 Tryptophan synthase beta chain (EC 4.2.1.20) | Rv1613 Tryptophan synthase alpha chain (EC 4.2.1.20) |
| Rv2127 L-asparagine permease | Rv2128 Possible membrane protein |
| Rv0176 FIG033285: Conserved MCE associated transmembrane protein | Rv0247c Succinate dehydrogenase iron-sulfur protein (EC 1.3.99.1) |
| Rv1614 Prolipoprotein diacylglyceryl transferase (EC 2.4.99.-) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006568 | GO:0006586 | GO:0042430 | GO:0006576 | GO:0022804 | GO:0005215 | tryptophan metabolic process | indolalkylamine metabolic process | indole-containing compound metabolic pro... | cellular biogenic amine metabolic proces... | active transmembrane transporter activit... | transporter activity |
|
GO:0005886 | plasma membrane |
|
GO:0044117 | growth of symbiont in host |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0005887 | integral to plasma membrane |
|
GO:0052572 | response to host immune response |
|
GO:0016887 | ATPase activity |
|
GO:0005524 | ATP binding |
|
GO:0005515 | protein binding |
|
GO:0005576 | extracellular region |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0006817 | phosphate ion transport |
|
GO:0016036 | cellular response to phosphate starvation |
|
GO:0042301 | phosphate ion binding |
|
GO:0005215 | transporter activity |
|
GO:0016020 | membrane |
|
GO:0006810 | transport |
|
GO:0004834 | tryptophan synthase activity |
|
GO:0040007 | growth |
|
GO:0000104 | succinate dehydrogenase activity |

















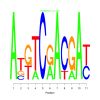
Discussion