Module 271 Residual: 0.56
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0271 | v02 | 0.56 | -22.32 |
| Rv1652 N-acetyl-gamma-glutamyl-phosphate reductase (EC 1.2.1.38) | Rv1653 Glutamate N-acetyltransferase (EC 2.3.1.35) / N-acetylglutamate synthase (EC 2.3.1.1) |
| Rv1654 Acetylglutamate kinase (EC 2.7.2.8) | Rv1655 Acetylornithine aminotransferase (EC 2.6.1.11) |
| Rv1656 Ornithine carbamoyltransferase (EC 2.1.3.3) | Rv1658 Argininosuccinate synthase (EC 6.3.4.5) |
| Rv1659 Argininosuccinate lyase (EC 4.3.2.1) | Rv2403c Lipoprotein LppR |
| Rv2404c Translation elongation factor LepA | Rv2783c Polyribonucleotide nucleotidyltransferase (EC 2.7.7.8) |
| Rv2796c PROBABLE CONSERVED LIPOPROTEIN LPPV | Rv2798c |
| Rv2799 Possible membrane protein | Rv2800 Possible hydrolase |
| Rv2703 RNA polymerase sigma factor SigB | Rv2782c FIG007959: peptidase, M16 family |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0046394 | GO:0016053 | carboxylic acid biosynthetic process | organic acid biosynthetic process |
|
GO:0003942 | N-acetyl-gamma-glutamyl-phosphate reductase activity |
|
GO:0040007 | growth |
|
GO:0042803 | protein homodimerization activity |
|
GO:0003991 | acetylglutamate kinase activity |
|
GO:0005886 | plasma membrane |
|
GO:0004585 | ornithine carbamoyltransferase activity |
|
GO:0006526 | arginine biosynthetic process |
|
GO:0070207 | protein homotrimerization |
|
GO:0004055 | argininosuccinate synthase activity |
|
GO:0005829 | cytosol |
|
GO:0004056 | argininosuccinate lyase activity |
|
GO:0006412 | translation |
|
GO:0005525 | GTP binding |
|
GO:0005618 | cell wall |
|
GO:0004654 | polyribonucleotide nucleotidyltransferase activity |
|
GO:0003700 | sequence-specific DNA binding transcription factor activity |
|
GO:0006355 | regulation of transcription, DNA-dependent |
|
GO:0016987 | sigma factor activity |
|
GO:0030528 | transcription regulator activity |
|
GO:0003677 | DNA binding |
|
GO:0006352 | DNA-dependent transcription, initiation |
|
GO:0005515 | protein binding |
|
GO:0009405 | pathogenesis |
|
GO:0009415 | response to water stimulus |
|
GO:0052572 | response to host immune response |


















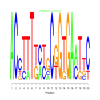
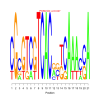
Discussion