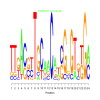
Module 298 Residual: 0.51
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0298 | v02 | 0.51 | -17.44 |
| Rv0913c Lignostilbene-alpha,beta-dioxygenase and related enzymes | Rv0914c 3-ketoacyl-CoA thiolase (EC 2.3.1.16) |
| Rv1880c Cytochrome P450 140 | Rv2338c Molybdopterin biosynthesis protein MoeW |
| Rv3057c Short chain dehydrogenase | Rv3058c Transcriptional regulator, TetR family |
| Rv3059 Cytochrome P450 136 | Rv3733c MutT-like protein |
| Rv3734c Wax ester synthase/acyl-CoA:diacylglycerol acyltransferase | Rv3735 Probable transmembrane protein |
| Rv0120c Translation elongation factor G-related protein | Rv1269c Possible membrane protein |
| Rv1270c Lipoprotein LprA | Rv1743 Serine/threonine-protein kinase PknE (EC 2.7.11.1) |
| Rv1829 | Rv1878 Glutamine synthetase (EC 6.3.1.2), putative |
| Rv1879 Amidohydrolase |
|
GO:0040007 | growth |
|
GO:0005886 | plasma membrane |
|
GO:0001666 | response to hypoxia |
|
GO:0004144 | diacylglycerol O-acyltransferase activity |
|
GO:0045017 | glycerolipid biosynthetic process |
|
GO:0071731 | response to nitric oxide |
|
GO:0005525 | GTP binding |
|
GO:0006412 | translation |
|
GO:0005618 | cell wall |
|
GO:0005576 | extracellular region |
|
GO:0042785 | active evasion of host immune response via regulation of host cytokine network |
|
GO:0048018 | receptor agonist activity |
|
GO:0004672 | protein kinase activity |
|
GO:0004674 | protein serine/threonine kinase activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0043085 | positive regulation of catalytic activity |
|
GO:0043086 | negative regulation of catalytic activity |
|
GO:0046777 | protein autophosphorylation |
|
GO:0051409 | response to nitrosative stress |
|
GO:0004356 | glutamate-ammonia ligase activity |
|
GO:0051260 | protein homooligomerization |



















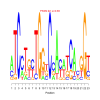
Discussion