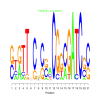
Module 326 Residual: 0.49
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0326 | v02 | 0.49 | -24.78 |
| Rv0975c Isovaleryl-CoA dehydrogenase (EC 1.3.99.10) | Rv0261c Nitrate/nitrite transporter |
| Rv0971c Methylglutaconyl-CoA hydratase (EC 4.2.1.18) | Rv0974c Methylcrotonyl-CoA carboxylase carboxyl transferase subunit (EC 6.4.1.4) |
| Rv0976c expressed protein | Rv0977 PE-PGRS FAMILY PROTEIN |
| Rv1575 | Rv1868 Phosphoenolpyruvate synthase (EC 2.7.9.2) |
| Rv1888c Possible membrane protein | Rv1999c Uncharacterized transporter Rv1999c/MT2055 |
| Rv2037c lysophospholipase-like family protein | Rv2040c Probable sugar-transport integral membrane protein ABC transporter |
| Rv2041c Probable sugar-binding lipoprotein | Rv2042c Putative uncharacterized protein BCG_2061c |
| Rv2043c Nicotinamidase (EC 3.5.1.19) | Rv2285 Diacyglycerol O-acyltransferase (EC 2.3.1.20) |
|
GO:0008470 | isovaleryl-CoA dehydrogenase activity |
|
GO:0004490 | methylglutaconyl-CoA hydratase activity |
|
GO:0005886 | plasma membrane |
|
GO:0004485 | methylcrotonoyl-CoA carboxylase activity |
|
GO:0071768 | mycolic acid biosynthetic process |
|
GO:0005829 | cytosol |
|
GO:0008986 | pyruvate, water dikinase activity |
|
GO:0016020 | membrane |
|
GO:0006810 | transport |
|
GO:0005215 | transporter activity |
|
GO:0040007 | growth |
|
GO:0008936 | nicotinamidase activity |
|
GO:0005506 | iron ion binding |
|
GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides |
|
GO:0030145 | manganese ion binding |
|
GO:0004144 | diacylglycerol O-acyltransferase activity |
|
GO:0045017 | glycerolipid biosynthetic process |


















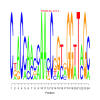
Discussion