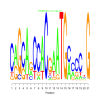
Module 367 Residual: 0.59
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0367 | v02 | 0.59 | -19.12 |
| Rv1093 Serine hydroxymethyltransferase 1 (EC 2.1.2.1) | Rv2840c COG2740: Predicted nucleic-acid-binding protein implicated in transcription termination |
| Rv2841c Transcription termination protein NusA | Rv2842c COG0779: clustered with transcription termination protein NusA |
| Rv0002 DNA polymerase III beta subunit (EC 2.7.7.7) | Rv0003 DNA recombination and repair protein RecF |
| Rv0237 Beta-hexosaminidase (EC 3.2.1.52) | Rv2274c |
| Rv2275 | Rv2450c Putative saccharopine dehydrogenase |
| Rv2451 | Rv2843 PROBABLE CONSERVED TRANSMEMBRANE ALANINE RICH PROTEIN |
| Rv2844 | Rv3434c POSSIBLE CONSERVED TRANSMEMBRANE PROTEIN |
| Rv3435c PROBABLE CONSERVED TRANSMEMBRANE PROTEIN | Rv3782 Putative glycosyl transferase |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006457 | GO:0004658 | protein folding | propionyl-CoA carboxylase activity |
|
GO:0004372 | glycine hydroxymethyltransferase activity |
|
GO:0005886 | plasma membrane |
|
GO:0006544 | glycine metabolic process |
|
GO:0006563 | L-serine metabolic process |
|
GO:0030170 | pyridoxal phosphate binding |
|
GO:0042783 | active evasion of host immune response |
|
GO:0042803 | protein homodimerization activity |
|
GO:0003723 | RNA binding |
|
GO:0003676 | nucleic acid binding |
|
GO:0030401 | transcription antiterminator activity |
|
GO:0031564 | transcription antitermination |
|
GO:0040007 | growth |
|
GO:0003887 | DNA-directed DNA polymerase activity |
|
GO:0005576 | extracellular region |
|
GO:0005829 | cytosol |
|
GO:0005618 | cell wall |
|
GO:0040008 | regulation of growth |
|
GO:0010628 | positive regulation of gene expression |
|
GO:0010629 | negative regulation of gene expression |
|
GO:0040010 | positive regulation of growth rate |
|
GO:0008378 | galactosyltransferase activity |


















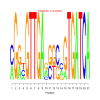
Discussion