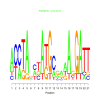
Module 402 Residual: 0.39
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0402 | v02 | 0.39 | -17.51 |
| Rv0047c Transcriptional regulator, PadR family | Rv0350 Chaperone protein DnaK |
| Rv0351 Heat shock protein GrpE | Rv0352 Chaperone protein DnaJ |
| Rv0353 HspR, transcriptional repressor of DnaK operon | Rv0814c probable sseC protein |
| Rv0815c Thiosulfate sulfurtransferase, rhodanese (EC 2.8.1.1) | Rv0859 3-ketoacyl-CoA thiolase [isoleucine degradation] (EC 2.3.1.16) |
| Rv0860 Enoyl-CoA hydratase [isoleucine degradation] (EC 4.2.1.17) / 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35) / 3-hydroxybutyryl-CoA epimerase (EC 5.1.2.3) | Rv1636 Universal stress protein UspA and related nucleotide-binding proteins |
| Rv3117 Thiosulfate sulfurtransferase, rhodanese (EC 2.8.1.1) | Rv3118 probable sseC protein |
| Rv3219 WhiB-like transcription regulator | Rv0046c Inositol-1-phosphate synthase (EC 5.5.1.4) |
| Rv3801c Long-chain fatty-acid-AMP ligase, Mycobacterial subgroup FadD32 |
|
GO:0005576 | extracellular region |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0009408 | response to heat |
|
GO:0040007 | growth |
|
GO:0046688 | response to copper ion |
|
GO:0046777 | protein autophosphorylation |
|
GO:0071451 | cellular response to superoxide |
|
GO:0051087 | chaperone binding |
|
GO:0000774 | adenyl-nucleotide exchange factor activity |
|
GO:0006457 | protein folding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0009267 | cellular response to starvation |
|
GO:0051082 | unfolded protein binding |
|
GO:0031072 | heat shock protein binding |
|
GO:0010468 | regulation of gene expression |
|
GO:0045892 | negative regulation of transcription, DNA-dependent |
|
GO:0003988 | acetyl-CoA C-acyltransferase activity |
|
GO:0005515 | protein binding |
|
GO:0015035 | protein disulfide oxidoreductase activity |
|
GO:0047134 | protein-disulfide reductase activity |
|
GO:0004512 | inositol-3-phosphate synthase activity |
|
GO:0008270 | zinc ion binding |
|
GO:0009405 | pathogenesis |
|
GO:0004467 | long-chain fatty acid-CoA ligase activity |
|
GO:0070566 | adenylyltransferase activity |
|
GO:0071769 | mycolate cell wall layer assembly |

















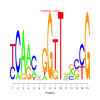
Discussion