Module 404 Residual: 0.54
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0404 | v02 | 0.54 | -5.82 |
| Rv0306 Cobalamin biosynthesis protein BluB @ 5,6-dimethylbenzimidazole synthase, flavin destructase family | Rv0571c Phosphoribosyl transferase domain protein |
| Rv1373 Glycolipid sulfotransferase | Rv1490 Possible membrane protein |
| Rv2028c Universal stress protein family | Rv2669 |
| Rv3079c Probable F420-dependent oxidoreductase family protein | Rv0570 Ribonucleotide reductase of class II (coenzyme B12-dependent) (EC 1.17.4.1) |
| Rv1039c PPE family protein | Rv1040c PPE family protein |
| Rv1557 Transmembrane transport protein MmpL6 | Rv1997 Probable cation-transporting ATPase F (EC 3.6.3.-) |
| Rv2003c | Rv2026c Universal stress protein family |
| Rv2029c Tagatose-6-phosphate kinase (EC 2.7.1.144) / 1-phosphofructokinase (EC 2.7.1.56) | Rv2624c Universal stress protein family |
| Rv2625c Probable conserved transmembrane alanine and leucine rich protein | Rv2666 Mobile element protein |
| Rv3468c UDP-glucose 4-epimerase (EC 5.1.3.2) |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0008146 | sulfotransferase activity |
|
GO:0040007 | growth |
|
GO:0005886 | plasma membrane |
|
GO:0071500 | cellular response to nitrosative stress |
|
GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor |
|
GO:0010033 | response to organic substance |
|
GO:0005887 | integral to plasma membrane |





















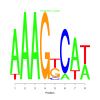

Discussion