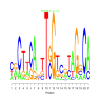
Module 429 Residual: 0.52
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0429 | v02 | 0.52 | -11.70 |
| Rv0620 Galactokinase (EC 2.7.1.6) | Rv0979c |
| Rv1271c | Rv1549 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) |
| Rv1550 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) | Rv1551 Glycerol-3-phosphate acyltransferase (EC 2.3.1.15) |
| Rv1553 Succinate dehydrogenase iron-sulfur protein (EC 1.3.99.1) | Rv1620c Transport ATP-binding protein CydC |
| Rv2491 Rossmann-fold nucleotide-binding protein | Rv2730 |
| Rv3273 Carbonic anhydrase (EC 4.2.1.1) | Rv0311 |
| Rv0618 Galactose-1-phosphate uridylyltransferase (EC 2.7.7.10) | Rv0891c Adenylyl cyclase class-3/4/guanylyl cyclase |
| Rv1469 Lead, cadmium, zinc and mercury transporting ATPase (EC 3.6.3.3) (EC 3.6.3.5); Copper-translocating P-type ATPase (EC 3.6.3.4) | Rv1552 Succinate dehydrogenase flavoprotein subunit (EC 1.3.99.1) |
| Rv1554 Fumarate reductase subunit C | Rv1555 Fumarate reductase subunit D |
| Rv3359 NADH:flavin oxidoreductases, Old Yellow Enzyme family |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0016741 | transferase activity, transferring one-c... |
|
GO:0004335 | galactokinase activity |
|
GO:0005618 | cell wall |
|
GO:0005886 | plasma membrane |
|
GO:0004366 | glycerol-3-phosphate O-acyltransferase activity |
|
GO:0000104 | succinate dehydrogenase activity |
|
GO:0005524 | ATP binding |
|
GO:0016021 | integral to membrane |
|
GO:0006810 | transport |
|
GO:0016887 | ATPase activity |
|
GO:0004089 | carbonate dehydratase activity |
|
GO:0005887 | integral to plasma membrane |
|
GO:0009405 | pathogenesis |
|
GO:0040007 | growth |
|
GO:0017103 | UTP:galactose-1-phosphate uridylyltransferase activity |
|
GO:0005829 | cytosol |
|
GO:0016020 | membrane |
|
GO:0006118 | electron transport |
|
GO:0006106 | fumarate metabolic process |





















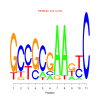
Discussion