Module 473 Residual: 0.49
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0473 | v02 | 0.49 | -34.36 |
| Rv0505c Phosphoserine phosphatase (EC 3.1.3.3) | Rv0506 membrane protein, MmpS family |
| Rv1326c 1,4-alpha-glucan (glycogen) branching enzyme, GH-13-type (EC 2.4.1.18) | Rv1327c Putative glucanase glgE (EC 3.2.1.-) |
| Rv1328 Glycogen phosphorylase (EC 2.4.1.1) | Rv0507 Putative membrane protein |
| Rv0508 | Rv0896 Citrate synthase (si) (EC 2.3.3.1) |
| Rv3205c |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0005515 | GO:0004180 | protein binding | carboxypeptidase activity |
|
GO:0005886 | plasma membrane |
|
GO:0005887 | integral to plasma membrane |
|
GO:0005576 | extracellular region |
|
GO:0003844 | 1,4-alpha-glucan branching enzyme activity |
|
GO:0005515 | protein binding |
|
GO:0005978 | glycogen biosynthetic process |
|
GO:0009250 | glucan biosynthetic process |
|
GO:0040007 | growth |
|
GO:0005829 | cytosol |
|
GO:0016758 | transferase activity, transferring hexosyl groups |
|
GO:0004645 | phosphorylase activity |
|
GO:0005618 | cell wall |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0004108 | citrate (Si)-synthase activity |











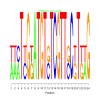
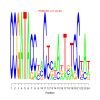
Discussion