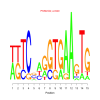
Module 486 Residual: 0.55
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0486 | v02 | 0.55 | -9.11 |
| Rv3416 Sporulation regulatory protein WhiD | Rv0112 GDP-mannose 4,6 dehydratase (EC 4.2.1.47) |
| Rv0113 Phosphoheptose isomerase (EC 5.3.1.-) | Rv0691c Mycofactocin system transcriptional regulator |
| Rv0726c | Rv1963c Transcriptional repressor Mce3R, TetR family |
| Rv2871 | Rv3016 Lipoprotein LpqA |
| Rv3033 | Rv3047c |
| Rv3048c Ribonucleotide reductase of class Ib (aerobic), beta subunit (EC 1.17.4.1) | Rv3451 serine esterase, cutinase family |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0003677 | GO:0003676 | DNA binding | nucleic acid binding |
|
GO:0005515 | protein binding |
|
GO:0015035 | protein disulfide oxidoreductase activity |
|
GO:0051536 | iron-sulfur cluster binding |
|
GO:0051539 | 4 iron, 4 sulfur cluster binding |
|
GO:0040007 | growth |
|
GO:0005975 | carbohydrate metabolic process |
|
GO:0005529 | NA |
|
GO:0003700 | sequence-specific DNA binding transcription factor activity |
|
GO:0045892 | negative regulation of transcription, DNA-dependent |
|
GO:0005886 | plasma membrane |
|
GO:0045927 | positive regulation of growth |
|
GO:0005576 | extracellular region |
|
GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor |
|
GO:0005971 | ribonucleoside-diphosphate reductase complex |
|
GO:0009263 | deoxyribonucleotide biosynthetic process |














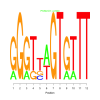
Discussion