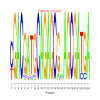
Module 496 Residual: 0.57
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0496 | v02 | 0.57 | -22.08 |
| Rv2259 Formaldehyde dehydrogenase MscR, NAD/mycothiol-dependent (EC 1.2.1.66) / S-nitrosomycothiol reductase MscR | Rv2260 Putative hydrolase in cluster with formaldehyde/S-nitrosomycothiol reductase MscR |
| Rv2680 Uncharacterized protein Q1 colocalized with Q | Rv2882c Ribosome recycling factor |
| Rv0491 Phosphate regulon transcriptional regulatory protein PhoB (SphR); Sensory transduction protein regX3 | Rv0527 Cytochrome c-type biogenesis protein CcdA (DsbD analog) |
| Rv0528 Ccs1/ResB-related putative cytochrome C-type biogenesis protein | Rv1341 Nucleoside 5-triphosphatase RdgB (dHAPTP, dITP, XTP-specific) (EC 3.6.1.15) |
| Rv1445c 6-phosphogluconolactonase (EC 3.1.1.31), eukaryotic type | Rv1446c OpcA, an allosteric effector of glucose-6-phosphate dehydrogenase, actinobacterial |
| Rv1447c Glucose-6-phosphate 1-dehydrogenase (EC 1.1.1.49) | Rv1449c Transketolase (EC 2.2.1.1) |
| Rv1457c \ABC-type multidrug transport system, permease component\\\"" | Rv1458c ABC-type multidrug transport system, ATPase component |
| Rv2257c Beta-lactamase class C and other penicillin binding proteins | Rv2258c Possible transcriptional regulatory protein |
| Rv2681 Ribonuclease D (EC 3.1.26.3) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006790 | sulfur compound metabolic process |
|
GO:0010127 | mycothiol-dependent detoxification |
|
GO:0016491 | oxidoreductase activity |
|
GO:0050607 | mycothiol-dependent formaldehyde dehydrogenase activity |
|
GO:0055114 | oxidation-reduction process |
|
GO:0004416 | hydroxyacylglutathione hydrolase activity |
|
GO:0040007 | growth |
|
GO:0006412 | translation |
|
GO:0005886 | plasma membrane |
|
GO:0006415 | translational termination |
|
GO:0008079 | translation termination factor activity |
|
GO:0008150 | biological_process |
|
GO:0003677 | DNA binding |
|
GO:0009405 | pathogenesis |
|
GO:0006355 | regulation of transcription, DNA-dependent |
|
GO:0016020 | membrane |
|
GO:0017004 | cytochrome complex assembly |
|
GO:0017057 | 6-phosphogluconolactonase activity |
|
GO:0004345 | glucose-6-phosphate dehydrogenase activity |
|
GO:0004802 | transketolase activity |
|
GO:0005576 | extracellular region |
|
GO:0005829 | cytosol |
|
GO:0071456 | cellular response to hypoxia |
|
GO:0004525 | ribonuclease III activity |



















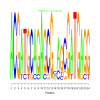
Discussion