Module 508 Residual: 0.53
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0508 | v02 | 0.53 | -27.36 |
| Rv1070c 3-methylthioacryloyl-CoA hydratase 2 (DmdD2) | Rv1071c 3-hydroxyisobutyryl-CoA hydrolase (EC 3.1.2.4) |
| Rv0066c Isocitrate dehydrogenase [NADP] (EC 1.1.1.42); Monomeric isocitrate dehydrogenase [NADP] (EC 1.1.1.42) | Rv1069c Predicted membrane protein (DUF2319) |
| Rv1072 PROBABLE CONSERVED TRANSMEMBRANE PROTEIN | Rv1326c 1,4-alpha-glucan (glycogen) branching enzyme, GH-13-type (EC 2.4.1.18) |
| Rv1327c Putative glucanase glgE (EC 3.2.1.-) | Rv1328 Glycogen phosphorylase (EC 2.4.1.1) |
| Rv2606c Pyridoxine biosynthesis glutamine amidotransferase, synthase subunit (EC 2.4.2.-) | Rv2607 Pyridoxamine 5'-phosphate oxidase (EC 1.4.3.5) |
|
GO:0004300 | enoyl-CoA hydratase activity |
|
GO:0005886 | plasma membrane |
|
GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity |
|
GO:0005829 | cytosol |
|
GO:0000287 | magnesium ion binding |
|
GO:0004448 | isocitrate dehydrogenase activity |
|
GO:0005576 | extracellular region |
|
GO:0042803 | protein homodimerization activity |
|
GO:0005887 | integral to plasma membrane |
|
GO:0003844 | 1,4-alpha-glucan branching enzyme activity |
|
GO:0005515 | protein binding |
|
GO:0005978 | glycogen biosynthetic process |
|
GO:0009250 | glucan biosynthetic process |
|
GO:0040007 | growth |
|
GO:0016758 | transferase activity, transferring hexosyl groups |
|
GO:0004645 | phosphorylase activity |
|
GO:0003674 | molecular_function |
|
GO:0004733 | pyridoxamine-phosphate oxidase activity |












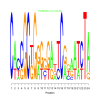
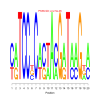
Discussion