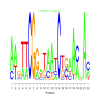
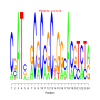
Module 552 Residual: 0.57
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0552 | v02 | 0.57 | -19.79 |
| Rv3355c | Rv3357 YefM protein (antitoxin to YoeB) |
| Rv3358 YoeB toxin protein | Rv3609c GTP cyclohydrolase I (EC 3.5.4.16) type 1 |
| Rv3610c Cell division protein FtsH (EC 3.4.24.-) | Rv2319c Universal stress protein family |
| Rv2322c Ornithine aminotransferase (EC 2.6.1.13) ; N-terminus domain | Rv2506 Transcriptional regulator, TetR family |
| Rv2572c Aspartyl-tRNA synthetase (EC 6.1.1.12) @ Aspartyl-tRNA(Asn) synthetase (EC 6.1.1.23) | Rv2573 2-dehydropantoate 2-reductase (EC 1.1.1.169) |
| Rv2779c Transcriptional regulator, AsnC family | Rv2966c Ribosomal RNA small subunit methyltransferase D (EC 2.1.1.-) |
| Rv3356c Methylenetetrahydrofolate dehydrogenase (NADP+) (EC 1.5.1.5) / Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9) | Rv3361c pentapeptide repeat family protein |
| Rv3364c | Rv3382c 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (EC 1.17.1.2) |
| Rv3383c Octaprenyl-diphosphate synthase (EC 2.5.1.-) / Dimethylallyltransferase (EC 2.5.1.1) / Geranyltranstransferase (farnesyldiphosphate synthase) (EC 2.5.1.10) / Geranylgeranyl pyrophosphate synthetase (EC 2.5.1.29) | Rv3611 |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006188 | GO:0006189 | GO:0046040 | IMP biosynthetic process | 'de novo' IMP biosynthetic process | IMP metabolic process |
|
GO:0040008 | regulation of growth |
|
GO:0003934 | GTP cyclohydrolase I activity |
|
GO:0040007 | growth |
|
GO:0008150 | biological_process |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0006508 | proteolysis |
|
GO:0030163 | protein catabolic process |
|
GO:0042783 | active evasion of host immune response |
|
GO:0005618 | cell wall |
|
GO:0008677 | 2-dehydropantoate 2-reductase activity |
|
GO:0005576 | extracellular region |
|
GO:0008657 | DNA topoisomerase (ATP-hydrolyzing) inhibitor activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0051745 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase activity |




















Discussion