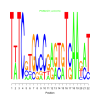
Module 560 Residual: 0.59
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0560 | v02 | 0.59 | -9.14 |
| Rv2482c Glycerol-3-phosphate acyltransferase (EC 2.3.1.15) | Rv2483c Phosphoserine phosphatase (EC 3.1.3.3) / 1-acyl-sn-glycerol-3-phosphate acyltransferase (EC 2.3.1.51) |
| Rv2484c Acyltransferase, ws/dgat/mgat subfamily protein | Rv0186 Beta-glucosidase (EC 3.2.1.21) |
| Rv0188 PROBABLE CONSERVED TRANSMEMBRANE PROTEIN | Rv0232 Transcriptional regulator, TetR family |
| Rv0233 Ribonucleotide-diphosphate reductase subunit beta (EC 1.17.4.1) | Rv0250c |
| Rv0275c Transcriptional regulator, TetR family | Rv0276 |
| Rv0670 Endonuclease IV (EC 3.1.21.2) | Rv1052 |
| Rv1264 Adenylate cyclase (EC 4.6.1.1) | Rv1316c Methylated-DNA--protein-cysteine methyltransferase (EC 2.1.1.63) |
| Rv1317c Methylated-DNA--protein-cysteine methyltransferase (EC 2.1.1.63) | Rv1454c Quinone oxidoreductase (EC 1.6.5.5) |
| Rv1996 Universal stress protein family | Rv2364c GTP-binding protein Era |
| Rv2557 | Rv3290c Probable L-lysine-epsilon aminotransferase (EC 2.6.1.36) (L-lysine aminotransferase) (Lysine 6-aminotransferase) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0009396 | GO:0042559 | GO:0006732 | GO:0042398 | folic acid-containing compound biosynthe... | pteridine-containing compound biosynthet... | coenzyme metabolic process | cellular modified amino acid biosyntheti... |
|
GO:0004366 | glycerol-3-phosphate O-acyltransferase activity |
|
GO:0005886 | plasma membrane |
|
GO:0005829 | cytosol |
|
GO:0008833 | deoxyribonuclease IV (phage-T4-induced) activity |
|
GO:0004016 | adenylate cyclase activity |
|
GO:0009268 | response to pH |
|
GO:0030145 | manganese ion binding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0009405 | pathogenesis |
|
GO:0003908 | methylated-DNA-[protein]-cysteine S-methyltransferase activity |
|
GO:0005618 | cell wall |
|
GO:0005576 | extracellular region |
|
GO:0071500 | cellular response to nitrosative stress |
|
GO:0003676 | nucleic acid binding |
|
GO:0005622 | intracellular |
|
GO:0005525 | GTP binding |
|
GO:0040007 | growth |
|
GO:0009267 | cellular response to starvation |






















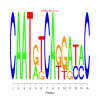
Discussion