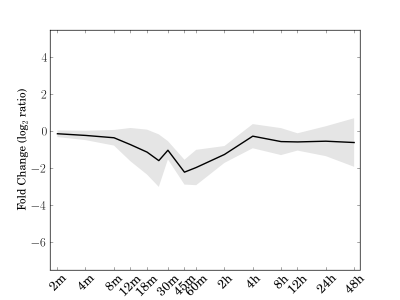
|
Cre01.g006100.t1.2
|
Cre01.g027450.t1.2 Thioredoxin superfamily protein
Details for Cre01.g027450.t1.2 - Thioredoxin superfamily protein
GO Terms: GO:0045454
Go to Gene Page: Cre01.g027450.t1.2
Phytozome Genomic Region

|
Cre01.g032650.t2.1 Aldolase superfamily protein
Details for Cre01.g032650.t2.1 - Aldolase superfamily protein
GO Terms: GO:0005975
Go to Gene Page: Cre01.g032650.t2.1
Phytozome Genomic Region

|
Cre01.g039750.t1.2 peptide deformylase 1B
Details for Cre01.g039750.t1.2 - peptide deformylase 1B
GO Terms: NA
Go to Gene Page: Cre01.g039750.t1.2
Phytozome Genomic Region

|
Cre02.g097100.t1.2 zinc finger (C3HC4-type RING finger) family protein
Details for Cre02.g097100.t1.2 - zinc finger (C3HC4-type RING finger) family protein
GO Terms: NA
Go to Gene Page: Cre02.g097100.t1.2
Phytozome Genomic Region

|
Cre03.g144947.t1.1 CCCH-type zinc finger protein with ARM repeat domain
Details for Cre03.g144947.t1.1 - CCCH-type zinc finger protein with ARM repeat domain
GO Terms: NA
Go to Gene Page: Cre03.g144947.t1.1
Phytozome Genomic Region

|
Cre03.g157564.t1.2
Details for Cre03.g157564.t1.2 -
GO Terms: NA
Go to Gene Page: Cre03.g157564.t1.2
Phytozome Genomic Region

|
Cre03.g159500.t5.1 Pyridoxal-dependent decarboxylase family protein
Details for Cre03.g159500.t5.1 - Pyridoxal-dependent decarboxylase family protein
GO Terms: GO:0003824
Go to Gene Page: Cre03.g159500.t5.1
Phytozome Genomic Region

|
Cre03.g163900.t1.1 Protein of unknown function (DUF506)
Details for Cre03.g163900.t1.1 - Protein of unknown function (DUF506)
GO Terms: NA
Go to Gene Page: Cre03.g163900.t1.1
Phytozome Genomic Region

|
Cre06.g271900.t2.1
Details for Cre06.g271900.t2.1 -
GO Terms: NA
Go to Gene Page: Cre06.g271900.t2.1
Phytozome Genomic Region

|
|
Cre06.g309450.t1.2 carboxypeptidase D, putative
|
Cre07.g348450.t1.1 calcium-dependent protein kinase 24
|
Cre08.g377850.t1.2
Details for Cre08.g377850.t1.2 -
GO Terms: NA
Go to Gene Page: Cre08.g377850.t1.2
Phytozome Genomic Region

|
Cre10.g424550.t1.1
Details for Cre10.g424550.t1.1 -
GO Terms: NA
Go to Gene Page: Cre10.g424550.t1.1
Phytozome Genomic Region

|
|
Cre10.g438850.t1.2 basic leucine-zipper 2
|
Cre12.g494400.t1.2 nucleotide binding protein 35
Details for Cre12.g494400.t1.2 - nucleotide binding protein 35
GO Terms: NA
Go to Gene Page: Cre12.g494400.t1.2
Phytozome Genomic Region

|
Cre12.g499352.t1.1
Details for Cre12.g499352.t1.1 -
GO Terms: NA
Go to Gene Page: Cre12.g499352.t1.1
Phytozome Genomic Region

|
Cre12.g533100.t1.1
Details for Cre12.g533100.t1.1 -
GO Terms: NA
Go to Gene Page: Cre12.g533100.t1.1
Phytozome Genomic Region

|
Cre12.g556150.t1.2 CAAX amino terminal protease family protein
Details for Cre12.g556150.t1.2 - CAAX amino terminal protease family protein
GO Terms: GO:0016020
Go to Gene Page: Cre12.g556150.t1.2
Phytozome Genomic Region

|
Cre14.g613200.t1.1 plastid transcriptionally active 17
Details for Cre14.g613200.t1.1 - plastid transcriptionally active 17
GO Terms: NA
Go to Gene Page: Cre14.g613200.t1.1
Phytozome Genomic Region

|
Cre14.g625650.t1.2
Details for Cre14.g625650.t1.2 -
GO Terms: NA
Go to Gene Page: Cre14.g625650.t1.2
Phytozome Genomic Region

|
Cre36.g759747.t1.1
Details for Cre36.g759747.t1.1 -
GO Terms: NA
Go to Gene Page: Cre36.g759747.t1.1
Phytozome Genomic Region

|

Comments