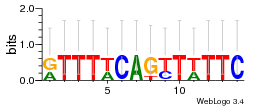
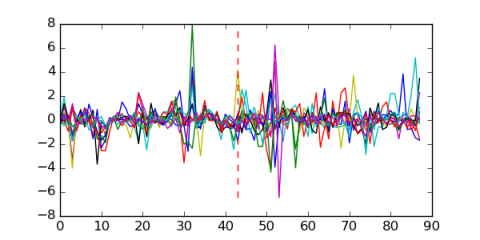
Module 201 Residual: 0.50
Corynebacterium glutamicum ATCC 13032
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.50 | 0.019 | 0.075 | 43 | 12 | 20160126 |
| cg2954 carbonic anhydrase (RefSeq) | cg0687 O-sialoglycoprotein endopeptidase (RefSeq) |
| cg2955 A/G-specific adenine glycosylase (RefSeq) | cg2590 xanthine/uracil permease (RefSeq) |
| cg0684 prolyl aminopeptidase A (RefSeq) | cg2589 GTPase ObgE (RefSeq) |
| cg0685 hypothetical protein (RefSeq) | cg2588 gamma-glutamyl kinase (RefSeq) |
| cg0686 GNAT family acetyltransferase (RefSeq) | cg2587 phosphoglycerate dehydrogenase or related dehydrogenase (RefSeq) |
| cg0683 hypothetical protein (RefSeq) | cg2586 gamma-glutamyl phosphate reductase (RefSeq) |
wildtype..FeSO4_vs_heme.3
GLY3_41_60min_rep1
Delta.chrSA_vs_WT_Fe_rep1
Delta.chrSA_vs_WT_Fe_rep2
strain.AR6.exponential.replica1
WT.30.mM.MeOH.vs.WT.2
Delta.chrSA_vs_WT_Heme_rep1
JVO1_vs_delta_fkpA_35degree_II
IPTG.concentration.of.10.uM
strain.AR1.exponential.replica2
wt_4glu_5_wt_2ac_3
LB.50mM.D.lactate_vs_LB.1
LB.50mM.D.lactate_vs_LB.2
tam45_41_0min_rep1
C..glutamicum.W.T_Furfural.6.5.mM_rep1
JVO1_vs_delta_fkpA_35degree_I
C..glutamicum.W.T_Furfural.6.5.mM_rep2
C..glutamicum.W.T_Furfural.20.mM_rep1
C..glutamicum.W.T_Furfural.20.mM_rep2
X21.25.uM.copper..wildtype_vs_Delta.copRS.rep2
X21.25.uM.copper..wildtype_vs_Delta.copRS.rep3
tam44_41_0min_rep1
X21.25.uM.copper..wildtype_vs_Delta.copRS.rep1
GLY3_41_30min_rep1
WT_vs_Delta.chrSA_Fe_rep3
WT_vs_DhrrA_heme.3
WT_vs_Delta.chrSA_Heme_rep3
strain.AR1.exponential.replica1
Glutamine_vs_Ammonia_rep1
LB_vs_LB.50mM.D.lactate
tam44_41_30min_rep1
WT.30.mM.MeOH.vs.WT.1
trm2_41_30min_rep1
C..glutamicum.W.T_Furfural.13.mM_rep2
C..glutamicum.W.T_Furfural.13.mM_rep1
WT_vs_DhrrA_FeSO4.1
Tol1.120.mM.MeOH.vs.Tol1.2
Tol1.120.mM.MeOH.vs.Tol1.1
wildtype..21.25.uM.copper_vs_1.25.uM.copper.rep1
rshA.mutant.at.the.exponential.phase_4th
tam44_41_15min_rep1
Ammonia_vs_Glutamine_rep2
Glutamine_vs_Ammonia_rep3
Comments
Log in to post comments