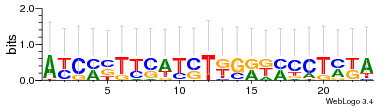
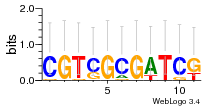
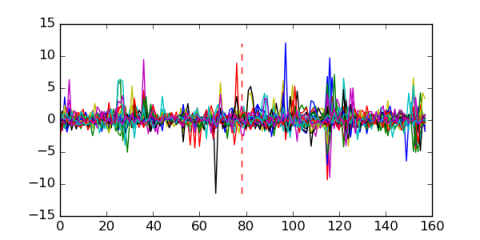
Module 2 Residual: 0.53
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.53 | 2.1e-28 | 0.083 | 78 | 33 | 20160204 |
| MSMEG_3079 Nucleotide-binding protein MSMEG_3079/MSMEI_3001 | MSMEG_3239 Putative signal transduction histidine kinase (Two-component system sensor kinase) |
| MSMEG_2490 Decarboxylase | MSMEG_4123 3-hydroxyisobutyrate dehydrogenase (6-phosphogluconate dehydrogenase NAD-binding (3-hydroxyisobutyrate dehydrogenase)) (EC 1.1.1.31) |
| MSMEG_4440 Glucose-1-dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) | MSMEG_3512 CinA-like protein |
| MSMEG_4441 Cupin domain protein | MSMEG_3472 Uncharacterized protein |
| MSMEG_3240 DNA-binding response regulator, LuxR family protein | MSMEG_3473 Uracil phosphoribosyltransferase (EC 2.4.2.9) (UMP pyrophosphorylase) (UPRTase) |
| MSMEG_2489 Transcriptional regulator, GntR family (Transcriptional regulator, GntR family protein, putative) | MSMEG_5709 Uncharacterized protein |
| MSMEG_4932 UDP-N-acetylglucosamine 1-carboxyvinyltransferase (EC 2.5.1.7) (Enoylpyruvate transferase) (UDP-N-acetylglucosamine enolpyruvyl transferase) | MSMEG_0736 Putative conserved secreted protein (Uncharacterized protein) |
| MSMEG_4758 Uncharacterized protein | MSMEG_5970 Glutamate-1-semialdehyde 2,1-aminomutase (EC 5.4.3.8) (Probable glutamate-1-semialdehyde 2,1-aminomutase) (EC 5.4.3.8) |
| MSMEG_0981 Two-component system regulator | MSMEG_3081 Putative sporulation transcription regulator WhiA |
| MSMEG_0980 Signal transduction histidine kinase (Two-component system sensor kinase) | MSMEG_3080 Putative gluconeogenesis factor |
| MSMEG_1761 Integral membrane protein | MSMEG_1758 Uncharacterized protein |
| MSMEG_6072 Arsenical pump membrane protein | MSMEG_3471 GTP cyclohydrolase (EC 3.5.4.25) |
| MSMEG_2653 Riboflavin biosynthesis protein (EC 2.7.1.26) (EC 2.7.7.2) | MSMEG_5969 Uncharacterized protein |
| MSMEG_2652 Iron (Metal) dependent repressor, DtxR family (Iron repressor protein) | MSMEG_0483 Major facilitator superfamily MFS_1 (Shikimate transporter) |
| MSMEG_1083 Uncharacterized protein | MSMEG_6581 Oxidoreductase, short chain dehydrogenase/reductase family protein (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) |
| MSMEG_0975 ATPases involved in chromosome partitioning (Uncharacterized protein) | MSMEG_2492 D-lactate dehydrogenase |
| MSMEG_4572 DNA polymerase III, delta subunit |
KO_stationary.phase_rep2
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
Wild.type.rep2.y
Biological.replicate.4.of.6..negative.control
Biological.replicate.5.of.7..panaxydol.treatment
M..smegmatis.Acid.NO.treated
Mycobacterium.smegmatis.Spheroplast.induced
Biological.replicate.2.of.6..isoniazid.treatment
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Wild.type.rep2.x
Wild.type.rep1.x
Wild.type.rep1.y
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X3dbfcy5plogcy3slide601norm
Sub.MIC.INH
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Slow.vs..Fast.Replicate.2
WT.vs..DamtR.N.surplus_rep1
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.1.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
X2.5..vs..50..air.saturation.Replicate.4
stphcy3plogcy5slide581norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
WT_stationary.phase_rep2
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.4
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
dosR.WT_Replicate3
Biological.replicate.4.of.7..falcarinol.treatment
Biological.replicate.6.of.7..ethambutol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
Wild.type.rep3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
Biological.replicate.2.of.6..negative.control
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
WT.vs..DamtR.N.surplus_rep2
dosR.WT_Replicate2
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments