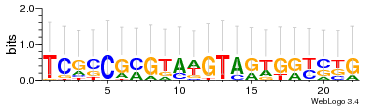
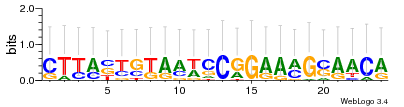
Module 8 Residual: 0.58
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.58 | 0.0000000000038 | 0.0000000000035 | 78 | 35 | 20160204 |
| MSMEG_6093 Uncharacterized protein | MSMEG_5150 Putative pterin-4-alpha-carbinolamine dehydratase (PHS) (EC 4.2.1.96) (4-alpha-hydroxy-tetrahydropterin dehydratase) (Pterin carbinolamine dehydratase) (PCD) |
| MSMEG_6091 ATP-dependent Clp protease ATP-binding subunit ClpC1 | MSMEG_2593 Gnat-family protein acetyltransferase |
| MSMEG_0965 Porin MspA | MSMEG_2639 Integral membrane protein |
| MSMEG_6157 DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I) | MSMEG_2804 Two-component system sensor kinase |
| MSMEG_3525 Transcriptional regulator, XRE family (Transcriptional regulator, XRE family protein) | MSMEG_2806 Two-component system response regulator |
| MSMEG_1595 Putative oxidoreductase (EC 1.-.-.-) (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) | MSMEG_5881 Carbon monoxide dehydrogenase subunit G (Putative carbon monoxide dehydrogenase subunit G) |
| MSMEG_4059 Haloacid dehalogenase, type II (EC 3.8.1.2) | MSMEG_5880 Nicotine dehydrogenase (Xanthine dehydrogenase, molybdenum binding subunit apoprotein) (EC 1.17.1.4) |
| MSMEG_6135 Uncharacterized protein | MSMEG_5883 (2Fe-2S)-binding protein (EC 1.-.-.-) ([2Fe-2S] binding domain protein) |
| MSMEG_4393 Carboxyvinyl-carboxyphosphonate phosphorylmutase (EC 2.7.8.23) (Putative carboxyvinyl-carboxyphosphonate phosphorylmutase 1 (Carboxyphosphonoenolpyruvate phosphonomutase)) | MSMEG_1596 Transcriptional regulator (Transcriptional regulator, TetR family) |
| MSMEG_2576 Deoxyribodipyrimidine photo-lyase (EC 4.1.99.3) | MSMEG_5882 Aerobic-type carbon monoxide dehydrogenase protein, FAD-binding subunit (Carbon monoxide dehydrogenase medium chain) (EC 1.2.99.2) |
| MSMEG_2578 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXP reductoisomerase) (EC 1.1.1.267) (1-deoxyxylulose-5-phosphate reductoisomerase) (2-C-methyl-D-erythritol 4-phosphate synthase) | MSMEG_6935 N-acetylmuramoyl-L-alanine amidase (EC 3.5.1.28) |
| MSMEG_2579 Zinc metalloprotease Rip1 (EC 3.4.24.-) (Regulator of sigma KLM proteases) (S2P endopeptidase) (Site-2-type intramembrane protease) (site-2 protease Rip1) (S2P protease Rip1) | MSMEG_3487 Metal dependent phosphohydrolase (EC 4.2.1.69) (Metal-dependent phosphohydrolase) |
| MSMEG_0477 Uncharacterized protein | MSMEG_4594 Uncharacterized protein |
| MSMEG_5976 Epimerase/dehydratase (NAD dependent epimerase/dehydratase) (EC 5.1.3.2) | MSMEG_4195 Uncharacterized protein |
| MSMEG_5879 D-alanyl-D-alanine dipeptidase (D-Ala-D-Ala dipeptidase) (EC 3.4.13.22) | MSMEG_4196 Lipoprotein lppL (LppL protein) |
| MSMEG_6197 Diaminopimelate decarboxylase (EC 4.1.1.20) | MSMEG_1315 Transporter, small conductance mechanosensitive ion channel (MscS) family protein |
| MSMEG_3133 Uncharacterized protein | MSMEG_3488 Transcriptional regulator, AraC family protein |
| MSMEG_3940 Universal stress protein family protein (UspA) |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
dcpA.knockout.rep2
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Bedaquiline.vs..DMSO.30min
Biological.replicate.2.of.7..falcarinol.treatment
dosR.WT_Replicate3
KO_log.phase_rep2
KO_log.phase_rep1
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
Biological.replicate.6.of.7..ethambutol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
stphcy5plogcy3slide487norm
Biological.replicate.1.of.7..panaxydol.treatment
Biological.replicate.3.of.6..negative.control
Biological.replicate.5.of.7..panaxydol.treatment
X4dbfcy5plogcy3slide480norm
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
X3dbfcy5plogcy3slide603norm
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Biological.replicate.2.of.7..panaxydol.treatment
X4XR.rep2
X4XR.rep1
Wild.type.rep2.x
Slow.vs..Fast.Replicate.5
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_dyeswap_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_dye.swap_rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..panaxydol.treatment
Wild.type.rep3
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments