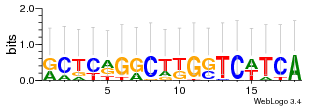
Module 11 Residual: 0.48
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.48 | 1.2e-35 | 0.0000000000039 | 77 | 29 | 20160204 |
| MSMEG_4628 Uncharacterized protein | MSMEG_5292 Uncharacterized protein |
| MSMEG_6882 LysR-family protein transcriptional regulator (Transcriptional regulator, LysR family) | MSMEG_5293 Uncharacterized protein |
| MSMEG_6884 NADP oxidoreductase coenzyme F420-dependent (NADP oxidoreductase, coenzyme f420-dependent:6-phosphogluconate dehydrogenase, nad-binding, putative) | MSMEG_1145 MCE-family protein Mce6C (Virulence factor Mce family protein) |
| MSMEG_1045 Integral membrane protein | MSMEG_1147 Mce family protein, Mce5E (Mce related protein) |
| MSMEG_1044 Uncharacterized protein | MSMEG_1143 MCE-family protein Mce6A (Mce related protein) |
| MSMEG_2273 [NiFe] hydrogenase maturation protein HypF | MSMEG_4065 Feruloyl-CoA synthetase |
| MSMEG_2272 Hydrogenase expression/synthesis, HypA (Hydrogenase nickel insertion protein HypA) | MSMEG_4023 Oxidoreductase (Pyridine nucleotide-disulfide oxidoreductase dimerization region) (EC 1.-.-.-) |
| MSMEG_2271 Hydrogenase accessory protein HypB | MSMEG_6643 DNA-binding protein (Transcriptional regulator, XRE family) |
| MSMEG_0470 Carboxylic ester hydrolase (EC 3.1.1.-) | MSMEG_6645 2-methylcitrate dehydratase 1 (EC 4.2.1.79) (2-methylcitrate dehydratase 2) |
| MSMEG_2276 Hydrogenase expression/formation protein HypE | MSMEG_2325 |
| MSMEG_2275 Hydrogenase expression/formation protein HypD | MSMEG_2324 |
| MSMEG_2274 Hydrogenase assembly chaperone HypC/HupF (Hydrogenase assembly chaperone hypC/hupF) | MSMEG_2323 |
| MSMEG_2690 DNA translocase FtsK | MSMEG_2115 Uncharacterized protein |
| MSMEG_1786 Stas domain, putative | MSMEG_2112 Secreted protein |
| MSMEG_5291 Acyl-CoA synthase |
dcpA.knockout.rep2
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
dosR.WT_Replicate3
dosR.WT_Replicate1
Biological.replicate.5.of.6..negative.control
stphcy5plogcy3slide487norm
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
DglnR_without.vs..with.N_rep1
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
DglnR_without.vs..with.N_rep2
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
X4XR.rep1
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
X0.6..vs..50..air.saturation.Replicate.3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
X0.6..vs..50..air.saturation.Replicate.4
Biological.replicate.2.of.7..falcarinol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
Plogcy5plogcy3slide113norm
mc2155.vs.Dcrp1.replicate.1
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
Biological.replicate.3.of.6..kanamycin.treatment
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X4dbfcy5plogcy3slide901norm
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X4dbfcy5plogcy3slide480norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
Wild.type.rep2.x
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.vs..DglnR.N_starvation_dye.swap_rep2
WT.vs..DamtR.N.surplus_dyeswap_rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments