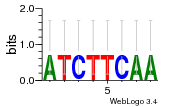
Module 12 Residual: 0.51
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.51 | 0.000000000000004 | 100000 | 77 | 23 | 20160204 |
| MSMEG_6713 Hydroxyquinol 1,2-dioxygenase (EC 1.13.11.-) (EC 1.13.11.37) | MSMEG_0498 Uncharacterized protein |
| MSMEG_4371 Putative iron-sulfur oxidoreductase (EC 1.14.12.7) (Vanillate O-demethylase oxidoreductase) (EC 1.14.13.-) | MSMEG_0112 Uncharacterized protein |
| MSMEG_4372 Capreomycidine hydroxylase (Rieske (2Fe-2S) region) | MSMEG_5051 Major facilitator superfamily MFS_1 (Major facilitator superfamily protein) |
| MSMEG_4607 Uncharacterized protein | MSMEG_1550 PduH protein (Uncharacterized protein) |
| MSMEG_2838 Uncharacterized protein | MSMEG_6417 Uncharacterized protein |
| MSMEG_0963 Uncharacterized protein | MSMEG_3475 Uncharacterized protein |
| MSMEG_5570 Multifunctional non-homologous end joining protein LigD (NHEJ DNA polymerase) [Includes: DNA repair polymerase (Pol) (Polymerase/primase); 3'-phosphoesterase (3'-ribonuclease/3'-phosphatase) (PE); DNA ligase (Lig) (EC 6.5.1.1) (Polydeoxyribonucleotide synthase [ATP])] | MSMEG_1678 LysR-family transcriptional regulator (Transcriptional regulator, LysR family protein) |
| MSMEG_1608 Glycosyl transferase | MSMEG_0636 Uncharacterized protein |
| MSMEG_3844 Lipid-transfer protein | MSMEG_1137 Amino acid permease-associated region |
| MSMEG_3843 Uncharacterized protein | MSMEG_3934 Phosphoenolpyruvate synthase (PEP synthase) (EC 2.7.9.2) (Pyruvate, water dikinase) |
| MSMEG_3453 Uncharacterized protein | MSMEG_1216 ABC-type transport system periplasmic substrate-binding protein |
| MSMEG_0499 Amino acid permease-associated region |
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
Wild.type.rep2.y
Biological.replicate.4.of.6..negative.control
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate2
Biological.replicate.4.of.6..kanamycin.treatment
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.2.of.7..falcarinol.treatment
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
Plogcy5plogcy3slide113norm
mc2155.vs.Dcrp1.replicate.1
X3dbfcy3plogcy5slide607norm
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_rep1
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
stphcy3plogcy5slide581norm
WT_log.phase_rep1
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide480norm
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
Biological.replicate.1.of.7..panaxydol.treatment
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_rep1
X2XR.rep1
Biological.replicate.7.of.7..panaxydol.treatment
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
Biological.replicate.2.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.1.of.6..kanamycin.treatment
Comments
Log in to post comments