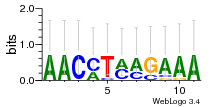
Module 22 Residual: 0.51
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.51 | 0.000000058 | 0.00046 | 73 | 33 | 20160204 |
| MSMEG_5484 Uncharacterized protein | MSMEG_4696 Alpha-amylase family protein (Putative alpha-glucosidase) (EC 3.2.1.20) |
| MSMEG_5124 2,4-dienoyl-coA reductase (Putative 2,4-dienoyl-CoA reductase NADPH) (EC 1.3.1.34) | MSMEG_6344 Transcriptional regulator |
| MSMEG_6673 6-aminohexanoate-cyclic-dimer hydrolase (EC 3.5.2.12) (Amidase) (EC 3.5.1.4) | MSMEG_0340 Enoyl-CoA hydratase/isomerase family protein |
| MSMEG_5504 Conserved secreted protein | MSMEG_2389 DNA-binding protein HU homolog (Histone-like protein) (Hlp) |
| MSMEG_0873 Uncharacterized protein | MSMEG_3977 Luciferase-like protein (EC 1.5.99.11) (Uncharacterized protein) |
| MSMEG_4191 Uncharacterized protein | MSMEG_3976 DNA-binding protein (Transcriptional regulator, TetR family) |
| MSMEG_5919 Uncharacterized protein | MSMEG_3583 Monooxygenase |
| MSMEG_5918 P450 heme-thiolate protein | MSMEG_0486 Putative ABC transporter, periplasmic protein |
| MSMEG_1104 Uncharacterized protein | MSMEG_0487 ABC transporter permease (Binding-protein-dependent transport systems inner membrane component) |
| MSMEG_2654 30S ribosomal protein S15 | MSMEG_4192 Uncharacterized protein |
| MSMEG_0011 FAD-binding 9, siderophore-interacting (Siderophore-interacting protein) | MSMEG_4334 Flavoprotein (Putative flavoprotein) (EC 2.5.1.26) |
| MSMEG_4335 Diacylglycerol kinase catalytic region (Uncharacterized protein) | MSMEG_4093 Oxidoreductase, aldo/keto reductase family (Oxidoreductase, aldo/keto reductase family protein) |
| MSMEG_5132 GTP-binding protein TypA/BipA (GTP-binding translation elongation factor TypA) (EC 3.6.5.3) | MSMEG_4091 Nitrilotriacetate monooxygenase component A (EC 1.14.13.-) |
| MSMEG_5135 Uncharacterized protein | MSMEG_2467 Branched-chain alpha-keto acid dehydrogenase E1 component (EC 1.2.4.4) (Transketolase, central region) |
| MSMEG_5887 Intersectin-EH binding protein Ibp1 | MSMEG_0488 ABC transporter ATP-binding protein |
| MSMEG_5886 Enoyl-CoA hydratase | MSMEG_4092 Alkanal monooxygenase (Luciferase family protein) (EC 1.14.14.3) |
| MSMEG_0366 Uncharacterized protein |
Wild.type.rep1.y
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Biological.replicate.5.of.7..panaxydol.treatment
X3dbfcy5plogcy3slide601norm
Biological.replicate.4.of.6..negative.control
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.1.of.6..negative.control
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
Biological.replicate.2.of.6..kanamycin.treatment
Wild.type.rep2.y
dosR.WT_Replicate4
Wild.type.rep1.x
DglnR_without.vs..with.N_rep1
DglnR_without.vs..with.N_rep2
MB100pruR.vs..MB100.rep.2
Biological.replicate.1.of.6..kanamycin.treatment
Bedaquiline.vs..DMSO.60min
Biological.replicate.6.of.6..kanamycin.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..isoniazid.treatment
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DamtR.N.surplus_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.2
Plogcy5plogcy3slide113norm
mc2155.vs.Dcrp1.replicate.1
mc2155.vs.Dcrp1.replicate.4
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
Biological.replicate.5.of.6..negative.control
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.1.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.4
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Slow.vs..Fast.Replicate.3
Biological.replicate.2.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.with.vs..without.N_rep2
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DglnR.N_starvation_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
dosR.WT_Replicate2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Comments
Log in to post comments