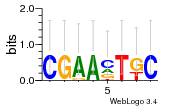
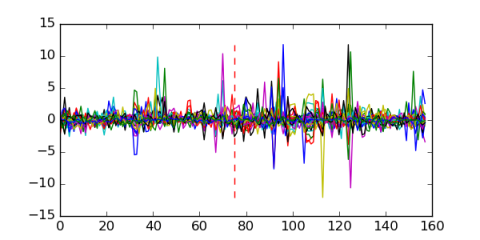
Module 46 Residual: 0.50
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.50 | 0.0002 | 0.47 | 75 | 30 | 20160204 |
| MSMEG_3726 Alcohol dehydrogenase (PQQ-dependent dehydrogenase, methanol/ethanol family) | MSMEG_4307 GTP cyclohydrolase 1 type 2 homolog |
| MSMEG_2188 Integral membrane protein | MSMEG_4306 Uncharacterized protein |
| MSMEG_3729 Catalase-peroxidase 3 (CP 3) (EC 1.11.1.21) (Peroxidase/catalase 3) | MSMEG_3611 Probable phosphoketolase (EC 4.1.2.-) |
| MSMEG_5247 PhoH family protein (PhoH-like protein phoH2 (Phosphate starvation-inducible protein psiH)) | MSMEG_3404 HNH endonuclease domain protein (Uncharacterized protein) |
| MSMEG_3461 Catalase-peroxidase 2 (CP 2) (EC 1.11.1.21) (Peroxidase/catalase 2) | MSMEG_6901 Uncharacterized protein |
| MSMEG_3460 Ferric uptake regulation protein (Ferric uptake regulator, Fur family) (EC 1.11.1.6) | MSMEG_6902 Putative membrane protein |
| MSMEG_3560 Glyoxalase/bleomycin resistance protein/dioxygenase (Uncharacterized protein) | MSMEG_5079 Uncharacterized protein |
| MSMEG_6468 Uncharacterized protein | MSMEG_5648 Uncharacterized protein |
| MSMEG_0412 PE (PE family protein) (Uncharacterized protein) | MSMEG_4702 ABC-type transporter, permease components |
| MSMEG_6232 Catalase (Catalase KatA) (EC 1.11.1.6) | MSMEG_1949 Uncharacterized protein |
| MSMEG_6234 CBS domain pair protein | MSMEG_3991 Cyclase (Putative cyclase) |
| MSMEG_2672 Acyl-CoA synthase (Fatty-acid-CoA ligase FadD23) | MSMEG_2010 Stress responsive A/B Barrel Domain superfamily protein (Stress responsive A/B barrel domain protein) |
| MSMEG_2671 Dihydrofolate reductase (EC 1.5.1.3) | MSMEG_6588 Fumarylacetoacetate |
| MSMEG_1089 Uncharacterized protein | MSMEG_1090 Amidase (EC 3.5.1.4) |
| MSMEG_4305 Phosphoglycerate mutase (Putative bifunctional ribonuclease H/phosphoglycerate mutase) (EC 3.1.26.4) | MSMEG_0387 Rmt2 protein |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
Wild.type.rep2.y
Bedaquiline.vs..DMSO.15min
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
Biological.replicate.2.of.6..isoniazid.treatment
Biological.replicate.4.of.7..ethambutol.treatment
Bedaquiline.vs..DMSO.30min
Biological.replicate.2.of.7..falcarinol.treatment
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate1
Biological.replicate.2.of.6..kanamycin.treatment
KO_log.phase_rep1
dosR.WT_Replicate4
Wild.type.rep1.x
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.7..ethambutol.treatment
DglnR_without.vs..with.N_rep2
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO..45min
MB100pruR.vs..MB100.rep.2
DamtR.with.vs..DamtR.without.N_.rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.3
Biological.replicate.1.of.7..panaxydol.treatment
Biological.replicate.3.of.6..negative.control
dcpA.knockout.rep2
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.N.surplus_rep1
X2.5..vs..50..air.saturation.Replicate.4
stphcy3plogcy5slide581norm
WT_log.phase_rep1
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.5.of.6..negative.control
mc2155.vs.Dcrp1.replicate.1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
Biological.replicate.2.of.6..negative.control
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments