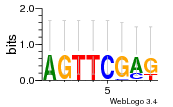
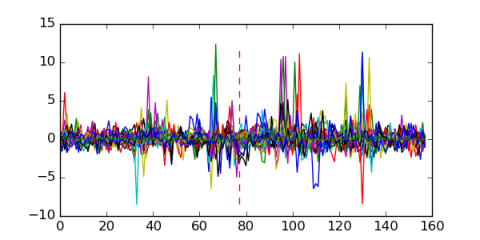
Module 49 Residual: 0.54
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.54 | 0.0000000000029 | 0.000012 | 77 | 37 | 20160204 |
| MSMEG_1915 Anti-sigma factor RshA (Regulator of SigH) (Sigma-H anti-sigma factor RshA) | MSMEG_4821 Uncharacterized protein |
| MSMEG_5774 tRNA-dihydrouridine synthase (EC 1.3.1.-) | MSMEG_4004 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100) |
| MSMEG_3342 Uncharacterized protein | MSMEG_4005 Calcium-binding protein (SMP-30/Gluconolaconase/LRE domain protein) (EC 3.1.1.17) |
| MSMEG_6158 Uncharacterized protein | MSMEG_2983 Uncharacterized protein |
| MSMEG_1868 Uncharacterized protein | MSMEG_0818 General substrate transporter (Transporter, major facilitator family protein) |
| MSMEG_1934 ATP-binding protein (Uncharacterized protein) | MSMEG_5825 Na+/H+ antiporter (NhaP-type Na+/H+ and K+/H+ antiporters with a unique C-terminal domain protein) |
| MSMEG_5465 Transcriptional regulator, AraC family protein | MSMEG_0320 Phosphotriesterase php (Parathion hydrolase) (EC 3.1.8.1) (Putative phosphotriesterase) |
| MSMEG_4549 Amino acid permease-associated region (Amino acid permease-associated region, putative) | MSMEG_3993 Asp/Glu racemase |
| MSMEG_1539 Uncharacterized protein | MSMEG_3994 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) |
| MSMEG_1538 Uncharacterized protein | MSMEG_1560 Uncharacterized protein |
| MSMEG_1458 Coenzyme PQQ synthesis C (EC 1.3.3.11) (Tena/thi-4 family protein) | MSMEG_0892 Conserved secreted protein (Uncharacterized protein) |
| MSMEG_1983 Major facilitator family protein transporter (Major facilitator superfamily MFS-1) | MSMEG_5926 Putative secreted protein |
| MSMEG_6057 Porin MspD | MSMEG_0897 Uncharacterized protein |
| MSMEG_0318 AMP-dependent synthetase and ligase | MSMEG_0896 Integral membrane transport protein (Major facilitator superfamily MFS_1) |
| MSMEG_6159 Probable cold shock protein A | MSMEG_3104 Uncharacterized protein |
| MSMEG_6598 6,7-dimethyl-8-ribityllumazine synthase (DMRL synthase) (LS) (Lumazine synthase) (EC 2.5.1.78) | MSMEG_3600 Uncharacterized protein |
| MSMEG_4550 Transposase | MSMEG_1212 Uncharacterized protein |
| MSMEG_6622 Cytochrome P450 (Cytochrome P450 monooxygenase) | MSMEG_1235 Sulfate permease (Sulfate transporter) |
| MSMEG_6531 Uncharacterized protein |
Wild.type.rep2.x
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
WT.vs..DamtR.without.N_rep2
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Wild.type.rep3
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate2
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.6.of.6..negative.control
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.6.of.7..ethambutol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
Wild.type.rep2.y
Biological.replicate.5.of.6..kanamycin.treatment
stphcy3plogcy5slide581norm
Biological.replicate.3.of.6..negative.control
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
Sub.MIC.INH
stphcy5plogcy3slide580norm
WT_log.phase_rep2
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X4XR.rep2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Biological.replicate.4.of.6..kanamycin.treatment
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments