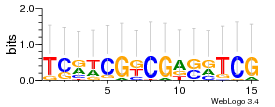
Module 50 Residual: 0.55
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.55 | 0.00000000000005 | 80 | 75 | 34 | 20160204 |
| MSMEG_3641 Uncharacterized protein (von Willebrand factor, type A) | MSMEG_2291 |
| MSMEG_2950 Carbon monoxide dehydrogenase medium chain (EC 1.2.99.2) (Carbon monoxide dehydrogenase, medium chain) | MSMEG_0171 Histone deacetylase superfamily protein |
| MSMEG_2951 (2Fe-2S)-binding protein (EC 1.2.99.2) ([2Fe-2S] binding domain protein) | MSMEG_6490 Major facilitator superfamily protein |
| MSMEG_5676 Citrate (Si) synthase (EC 2.3.3.1) (Citrate synthase) | MSMEG_2735 Diaminopimelate epimerase (DAP epimerase) (EC 5.1.1.7) |
| MSMEG_5677 Uncharacterized protein | MSMEG_2940 Probable transcriptional regulatory protein MSMEG_2940/MSMEI_2866 |
| MSMEG_5674 Major facilitator superfamily MFS_1 (Membrane transport protein) | MSMEG_4554 Probable serine/threonine-protein kinase PknH, putative (EC 2.7.11.1) (Serine/threonine protein kinase) |
| MSMEG_5675 Pyridoxine/pyridoxamine 5'-phosphate oxidase (EC 1.4.3.5) (PNP/PMP oxidase) (PNPOx) (Pyridoxal 5'-phosphate synthase) | MSMEG_2802 Uncharacterized protein |
| MSMEG_1492 MarR-transcriptional regulator (Transcriptional regulator, MarR family protein) | MSMEG_2949 Aldehyde oxidase and xanthine dehydrogenase molybdopterin binding protein (EC 1.17.1.4) (Carbon-monoxide dehydrogenase) |
| MSMEG_5735 Alpha-keto-acid decarboxylase (KDC) (EC 4.1.1.-) | MSMEG_6534 Uncharacterized protein |
| MSMEG_5734 Alpha-methylacyl-CoA racemase (Fatty-acid-CoA racemase Far) (EC 5.1.99.4) | MSMEG_5523 Peptidase (Peptidase S9, prolyl oligopeptidase active site region) (EC 3.4.19.1) |
| MSMEG_6506 Nicotinamidase/pyrazinamidase (Pyrazinamidase/nicotinamidase) (EC 3.5.1.19) | MSMEG_3532 Serine/threonine dehydratase family protein |
| MSMEG_5496 MscS Mechanosensitive ion channel | MSMEG_2482 Uncharacterized protein |
| MSMEG_1994 HpcH/HpaI aldolase | MSMEG_2033 NADPH quinone oxidoreductase FadB4 (EC 1.6.5.5) (Oxidoreductase, zinc-binding dehydrogenase family protein) |
| MSMEG_1993 MaoC like domain protein | MSMEG_0584 Uncharacterized protein |
| MSMEG_6373 Uncharacterized protein | MSMEG_0224 O-methyltransferase (O-methyltransferase MdmC) (EC 2.1.1.-) |
| MSMEG_2793 Sensor-type histidine kinase PrrB (EC 2.7.13.3) | MSMEG_5148 CTP pyrophosphohydrolase (EC 3.6.1.-) (Putative mutator protein MutT2/NUDIX hydrolase) |
| MSMEG_2414 Phosphopantetheine adenylyltransferase (EC 2.7.7.3) (Dephospho-CoA pyrophosphorylase) (Pantetheine-phosphate adenylyltransferase) (PPAT) | MSMEG_4314 Uncharacterized protein |
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Wild.type.rep2.y
WT.vs..DamtR.without.N_rep2
dosR.WT_Replicate4
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
mc2155.vs.Dcrp1.replicate.3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.15min
M..smegmatis.Acid.NO.treated
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.2.of.7..ethambutol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.2.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_rep1
Biological.replicate.5.of.6..isoniazid.treatment
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
DglnR_without.vs..with.N_rep1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Biological.replicate.2.of.6..kanamycin.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.3
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.vs..DamtR.N.surplus_dyeswap_rep2
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DglnR.N_starvation_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
dosR.WT_Replicate2
Biological.replicate.4.of.6..kanamycin.treatment
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments