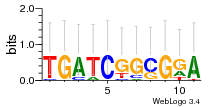
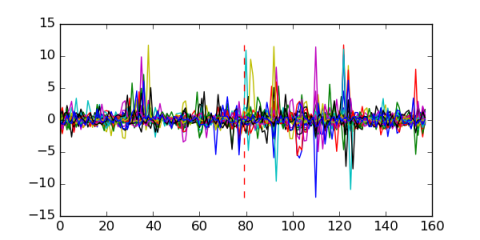
Module 72 Residual: 0.48
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.48 | 8.6e-25 | 0.48 | 79 | 36 | 20160204 |
| MSMEG_0742 LysR-family transcriptional regulator (Probable transcriptional regulatory protein) | MSMEG_5605 Cytochrome bd ubiquinol oxidase, subunit I |
| MSMEG_5635 Uncharacterized protein | MSMEG_4410 Uncharacterized protein |
| MSMEG_3223 TM2 domain containing protein+B7201 (TM2 domain protein) | MSMEG_1612 Extracellular solute-binding protein, family protein 3 |
| MSMEG_3224 Conserved membrane protein (Uncharacterized protein) | MSMEG_1613 ABC polar amino acid transporter, inner membrane subunit (Truncated polar amino acid ABC transporter) |
| MSMEG_4019 AMP-dependent synthetase and ligase | MSMEG_1614 Glutamine transport ATP-binding protein GlnQ |
| MSMEG_3467 Uncharacterized protein | MSMEG_3459 Uncharacterized protein |
| MSMEG_2148 HNH endonuclease domain protein | MSMEG_5375 GntR-family protein transcriptional regulator |
| MSMEG_0879 Uncharacterized protein | MSMEG_6189 Crp/Fnr familytranscriptional regulator (Transcriptional regulator, Crp/Fnr family protein) |
| MSMEG_0880 60 kDa chaperonin 1 (GroEL protein 1) (Protein Cpn60 1) | MSMEG_0563 Uncharacterized protein |
| MSMEG_0214 L-sorbosone dehydrogenase (SMP-30/Gluconolaconase/LRE-like region) | MSMEG_1379 Uncharacterized protein |
| MSMEG_3928 Nickel-dependent hydrogenase, large subunit (EC 1.12.-.-) ([NiFe] hydrogenase, alpha subunit, putative) | MSMEG_0992 2-nitropropane dioxygenase NPD (EC 1.13.12.16) (2-nitropropane dioxygenase, NPD) |
| MSMEG_3929 NADH ubiquinone oxidoreductase 20 kDa subunit ([NiFe] hydrogenase, delta subunit, putative) | MSMEG_3857 Uncharacterized protein |
| MSMEG_3926 ATPase, P-type (Transporting), HAD superfamily, subfamily IC (EC 3.6.3.8) (Cation-transporting ATPase Pma1) (EC 3.6.3.-) | MSMEG_3931 [NiFe] hydrogenase, beta subunit, putative |
| MSMEG_3927 Hydrogenase maturation protease (Peptidase M52, hydrogen uptake protein) | MSMEG_3668 Acyl-CoA dehydrogenase (Acyl-CoA dehydrogenase, type 2, C-terminal domain protein) |
| MSMEG_3858 RNA polymerase-binding protein RbpA | MSMEG_4253 GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase (EC 2.4.1.57) (Alpha-D-mannose-alpha-(1-6)-phosphatidylmyo-inositol-mannosyltransferase) (Alpha-mannosyltransferase) (Alpha-ManT) (Guanosine diphosphomannose-phosphatidyl-inositol alpha-mannosyltransferase) (Phosphatidylinositol alpha-mannosyltransferase) (PI alpha-mannosyltransferase) |
| MSMEG_3670 Small multidrug resistance protein (Transporter, small multidrug resistance (SMR) family protein) | MSMEG_3661 Uncharacterized protein |
| MSMEG_3672 Transporter, small multidrug resistance (SMR) family protein | MSMEG_3667 Carboxylic ester hydrolase (EC 3.1.1.-) |
| MSMEG_0851 Arsenic-transport integral membrane protein ArsA (Citrate transporter) | MSMEG_1213 Cytochrome P450 monooxygenase |
Wild.type.rep1.y
KO_stationary.phase_rep2
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
Biological.replicate.2.of.7..panaxydol.treatment
dcpA.knockout.rep2
Biological.replicate.4.of.6..negative.control
Bedaquiline.vs..DMSO.30min
Biological.replicate.3.of.7..ethambutol.treatment
MB100pruR.vs..MB100..rep.3
Biological.replicate.5.of.6..negative.control
KO_log.phase_rep2
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.3.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.3
stphcy3plogcy5slide581norm
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
WT_log.phase_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep1
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
mc2155.vs.Dcrp1.replicate.1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
stphcy5plogcy3slide487norm
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
Biological.replicate.1.of.6..negative.control
X2XR.rep2
Biological.replicate.5.of.7..falcarinol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DglnR.N_starvation_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.4.of.6..kanamycin.treatment
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments