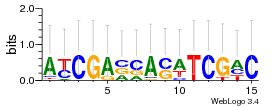
Module 73 Residual: 0.48
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.48 | 2e-29 | 0.0000019 | 78 | 33 | 20160204 |
| MSMEG_5105 Uncharacterized protein | MSMEG_3597 Uncharacterized protein |
| MSMEG_3339 Uncharacterized protein | MSMEG_2292 |
| MSMEG_6440 FAD-containing monooxygenase EthA (EC 1.14.13.-) (Baeyer-Villiger monooxygenase) (BVMO) (Prodrug activator EtaA) | MSMEG_5854 Transcriptional regulator, TetR family protein |
| MSMEG_6441 HTH-type transcriptional regulator EthR | MSMEG_4573 Competence protein (Uncharacterized protein) |
| MSMEG_1809 Sulfurtransferase | MSMEG_2787 Bifunctional enzyme riboflavin biosynthesis protein ribD : diaminohydroxyphosphoribosylaminopyrimidine deaminase + 5-amino-6-(5-phosphoribosylamino)uracil reductase (EC 1.1.1.193) (Putative riboflavin biosynthesis protein RibD) |
| MSMEG_1808 Fe-S metabolism associated SufE | MSMEG_1754 Uncharacterized protein |
| MSMEG_5349 Uncharacterized protein | MSMEG_5804 Uncharacterized protein |
| MSMEG_1603 Uncharacterized oxidoreductase MSMEG_1603/MSMEI_1564 (EC 1.-.-.-) | MSMEG_6245 Chloramphenicol exporter (Chloramphenicol resistance protein) |
| MSMEG_4739 Uncharacterized protein | MSMEG_0564 Xanthine/uracil permease (Xanthine/uracil/vitamin C permease) |
| MSMEG_1061 Phosphohydrolase (Pyridoxamine 5'-phosphate oxidase-related FMN-binding protein) | MSMEG_2997 Acyl-CoA thioesterase (Uncharacterized protein) |
| MSMEG_4526 Uncharacterized protein | MSMEG_2401 Uncharacterized protein |
| MSMEG_6196 Gaba permease | MSMEG_3973 N-methylhydantoinase (EC 3.5.2.9) |
| MSMEG_6194 Beta-lactamase related protein (Putative beta-lactamase) | MSMEG_1136 Uncharacterized protein |
| MSMEG_2619 Efflux protein (Integral membrane efflux protein EfpA) | MSMEG_4253 GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase (EC 2.4.1.57) (Alpha-D-mannose-alpha-(1-6)-phosphatidylmyo-inositol-mannosyltransferase) (Alpha-mannosyltransferase) (Alpha-ManT) (Guanosine diphosphomannose-phosphatidyl-inositol alpha-mannosyltransferase) (Phosphatidylinositol alpha-mannosyltransferase) (PI alpha-mannosyltransferase) |
| MSMEG_3598 ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein (EC 3.6.3.17) (Periplasmic sugar-binding proteins) | MSMEG_0223 Uncharacterized protein |
| MSMEG_1121 Uncharacterized protein | MSMEG_2441 Signal peptidase I (EC 3.4.21.89) |
| MSMEG_0222 Uncharacterized protein |
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Bedaquiline.vs..DMSO.15min
Biological.replicate.4.of.6..negative.control
Wild.type.rep2.x
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate3
dosR.WT_Replicate1
KO_log.phase_rep2
WT.vs..DamtR.N.surplus_dyeswap_rep2
Wild.type.rep1.x
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.2.of.7..falcarinol.treatment
Wild.type.rep3
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
M..smegmatis.Acid.NO.treated
WT.vs..DglnR.N_starvation_dye.swap_rep1
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DamtR.N.surplus_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.1.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
Biological.replicate.2.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
Biological.replicate.5.of.6..isoniazid.treatment
X2.5..vs..50..air.saturation.Replicate.4
Biological.replicate.4.of.7..ethambutol.treatment
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X2.5..vs..50..air.saturation.Replicate.2
Biological.replicate.2.of.6..negative.control
WT_stationary.phase_rep1
X4XR.rep2
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Bedaquiline.vs..DMSO.60min
Biological.replicate.2.of.6..kanamycin.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep2
X2XR.rep1
X2XR.rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments