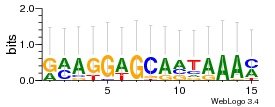
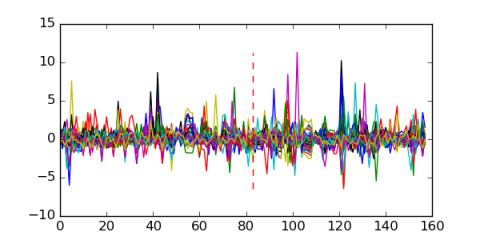
Module 79 Residual: 0.59
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.59 | 7.4e-27 | 0.000000000003 | 83 | 34 | 20160204 |
| MSMEG_5747 Uncharacterized protein | MSMEG_0855 Uncharacterized protein |
| MSMEG_4565 Aldo/keto reductase (EC 1.1.1.65) (Oxidoreductase) | MSMEG_1927 Cobyrinic Acid a,c-diamide synthase |
| MSMEG_6179 Acetyl-coenzyme A synthetase (AcCoA synthetase) (Acs) (EC 6.2.1.1) (Acetate--CoA ligase) (Acyl-activating enzyme) | MSMEG_6747 Oxidoreductase (Zinc-binding oxidoreductase) (EC 1.6.5.5) |
| MSMEG_3156 Uncharacterized protein | MSMEG_6746 Oxidoreductase, aldo/keto reductase family (Oxidoreductase, aldo/keto reductase family protein) |
| MSMEG_6464 Uncharacterized protein | MSMEG_4907 Mov34/MPN/PAD-1 (Mov34/MPN/PAD-1 family protein) |
| MSMEG_5837 Glutathione peroxidase | MSMEG_4906 ThiS family protein |
| MSMEG_4955 Homoserine kinase (HK) (HSK) (EC 2.7.1.39) | MSMEG_4905 Cysteine synthase B (EC 2.5.1.47) |
| MSMEG_4956 Threonine synthase (TS) (EC 4.2.3.1) | MSMEG_1776 Ribonuclease Z (RNase Z) (EC 3.1.26.11) (tRNA 3 endonuclease) (tRNase Z) |
| MSMEG_4957 Homoserine dehydrogenase (EC 1.1.1.3) | MSMEG_4124 Uncharacterized protein |
| MSMEG_4564 CBS domain protein (Putative signal-transduction protein with CBS domains) | MSMEG_3183 L-threonine dehydratase (EC 4.3.1.19) (Threonine deaminase) |
| MSMEG_4958 Diaminopimelate decarboxylase (DAP decarboxylase) (DAPDC) (EC 4.1.1.20) | MSMEG_1527 tRNA pseudouridine synthase A (EC 5.4.99.12) (tRNA pseudouridine(38-40) synthase) (tRNA pseudouridylate synthase I) (tRNA-uridine isomerase I) |
| MSMEG_4959 Arginine--tRNA ligase (EC 6.1.1.19) (Arginyl-tRNA synthetase) (ArgRS) | MSMEG_1524 DNA-directed RNA polymerase subunit alpha (RNAP subunit alpha) (EC 2.7.7.6) (RNA polymerase subunit alpha) (Transcriptase subunit alpha) |
| MSMEG_3065 Fmu (Sun) (Ribosomal RNA small subunit methyltransferase B) (EC 2.1.1.-) | MSMEG_1525 50S ribosomal protein L17 |
| MSMEG_4323 Pyruvate dehydrogenase E1 component (PDH E1 component) (EC 1.2.4.1) | MSMEG_0024 Peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) |
| MSMEG_4322 Uncharacterized protein | MSMEG_4271 Uncharacterized protein |
| MSMEG_3064 Methionyl-tRNA formyltransferase (EC 2.1.2.9) | MSMEG_4270 Adenosine kinase (Carbohydrate kinase CbhK) (EC 2.7.1.20) |
| MSMEG_2960 Preprotein translocase, YajC subunit (Protein translocase subunit yajC) | MSMEG_0023 Cell wall synthesis protein CwsA (Cell wall synthesis and cell shape protein A) |
mc2155.vs.Dcrp1.replicate.1
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
Biological.replicate.3.of.6..kanamycin.treatment
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
Mycobacterium.smegmatis.Spheroplast.induced
Plogcy5plogcy3slide110norm
Bedaquiline.vs..DMSO.30min
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
Biological.replicate.5.of.6..negative.control
Biological.replicate.2.of.6..kanamycin.treatment
Biological.replicate.7.of.7..falcarinol.treatment
X3dbfcy5plogcy3slide601norm
Wild.type.rep1.y
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.1.of.6..kanamycin.treatment
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.7..falcarinol.treatment
X4XR.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.4.of.6..kanamycin.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.5
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.5.of.6..kanamycin.treatment
dcpA.knockout.rep2
Biological.replicate.2.of.6..isoniazid.treatment
Biological.replicate.2.of.7..falcarinol.treatment
X3dbfcy5plogcy3slide603norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
X3dbfcy3plogcy5slide607norm
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.2.of.6..negative.control
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Bedaquiline.vs..DMSO.60min
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.3
Biological.replicate.4.of.6..negative.control
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
Wild.type.rep2.x
X2XR.rep2
WT.vs..DamtR.N.surplus_dyeswap_rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
Comments
Log in to post comments