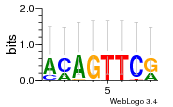
Module 109 Residual: 0.56
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.56 | 0.00000037 | 7.6 | 76 | 28 | 20160204 |
| MSMEG_3355 NmrA family protein (Uncharacterized protein) | MSMEG_6381 Uncharacterized protein |
| MSMEG_3016 Transglutaminase family protein (Uncharacterized protein) | MSMEG_3007 Succinate-semialdehyde dehydrogenase |
| MSMEG_3017 Uncharacterized protein | MSMEG_3006 Fe-dependent alcohol dehydrogenase (Iron-containing alcohol dehydrogenase) (EC 1.1.1.1) |
| MSMEG_0358 R2-like ligand binding oxidase (EC 1.-.-.-) (Ribonucleotide reductase R2 subunit homolog) (Ribonucleotide reductase small subunit homolog) | MSMEG_3005 Hydroxyacyl-CoA dehydrogenase |
| MSMEG_3462 Nitroreductase (Oxidoreductase) | MSMEG_6268 Putative ScnB protein |
| MSMEG_3018 Transglutaminase domain protein (Transglutaminase-like protein) | MSMEG_3381 FMN-dependent NADH-azoreductase (EC 1.7.-.-) (Azo-dye reductase) (FMN-dependent NADH-azo compound oxidoreductase) |
| MSMEG_3019 Uncharacterized protein | MSMEG_3382 ArsR family transcriptional regulator (Transcriptional regulator, ArsR family protein) |
| MSMEG_6271 2-isopropylmalate synthase (EC 2.3.3.13) (Alpha-IPM synthase) (Alpha-isopropylmalate synthase) | MSMEG_6266 Thiocyanate hydrolase beta subunit (EC 3.5.5.8) |
| MSMEG_4972 Acetyltransferase | MSMEG_6267 Thiocyanate hydrolase gamma subunit (EC 3.5.5.8) (Thiocyanate hydrolase subunit gamma) |
| MSMEG_4973 Glyoxalase/bleomycin resistance protein/dioxygenase | MSMEG_2389 DNA-binding protein HU homolog (Histone-like protein) (Hlp) |
| MSMEG_1348 RNA polymerase ECF-subfamily protein sigma factor (RNA polymerase sigma-70 factor, ECF subfamily) | MSMEG_5922 Lipid-transfer protein |
| MSMEG_1349 DGPFAETKE family protein (Dgpf domain family protein) | MSMEG_5923 Acetyl-CoA acetyltransferase (Lipid carrier protein or keto acyl-CoA thiolase Ltp3) (EC 2.3.1.-) |
| MSMEG_3008 Putative sigma 54 type regulator | MSMEG_5921 Uncharacterized protein |
| MSMEG_3672 Transporter, small multidrug resistance (SMR) family protein | MSMEG_6809 ATPase associated with various cellular activities AAA_5 (EC 1.7.2.5) (CbbQ protein) |
Wild.type.rep1.y
KO_stationary.phase_rep1
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
WT.vs..DamtR.without.N_rep2
M..smegmatis.Acid.NO.treated
Wild.type.rep2.x
Bedaquiline.vs..DMSO.30min
dosR.WT_Replicate2
dosR.WT_Replicate1
stphcy5plogcy3slide487norm
Wild.type.rep2.y
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
DglnR_without.vs..with.N_rep1
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Bedaquiline.vs..DMSO.60min
rel.knockout.rep2
Biological.replicate.1.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.2
Biological.replicate.6.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X0.6..vs..50..air.saturation.Replicate.5
Slow.vs..Fast.Replicate.2
Biological.replicate.2.of.7..falcarinol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.3
X3dbfcy3plogcy5slide607norm
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
Plogcy5plogcy3slide111norm
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy3plogcy5slide581norm
WT_log.phase_rep1
WT.with.vs..without.N_dye.swap_rep1
X4dbfcy5plogcy3slide480norm
Biological.replicate.2.of.7..ethambutol.treatment
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.1.of.7..ethambutol.treatment
X4XR.rep2
Biological.replicate.2.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
X3dbfcy5plogcy3slide603norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
X4dbfcy5plogcy3slide901norm
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
DglnR_without.vs..with.N_dyeswap_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.2.of.6..negative.control
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments