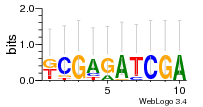
Module 110 Residual: 0.49
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.49 | 0.0000000000083 | 0.0000000037 | 77 | 37 | 20160204 |
| MSMEG_3282 Uncharacterized protein | MSMEG_5648 Uncharacterized protein |
| MSMEG_6467 DNA protection during starvation protein (EC 1.16.-.-) | MSMEG_6660 Permease, cytosine/purines, uracil, thiamine, allantoin family protein |
| MSMEG_5836 Uncharacterized protein | MSMEG_3531 Uncharacterized protein |
| MSMEG_2354 Nitroreductase (Oxidoreductase) | MSMEG_5231 Uncharacterized protein |
| MSMEG_6054 UPF0271 protein MSMEG_6054 | MSMEG_4925 Transcriptional regulator, Ada family protein/DNA-3-methyladenine glycosylase II (EC 3.2.2.21) |
| MSMEG_0884 Glyoxalase family protein (Glyoxalase/bleomycin resistance protein/dioxygenase) | MSMEG_0465 Transcriptional regulator, TetR family (Transcriptional regulator, TetR family protein) |
| MSMEG_1127 Probable conserved transmembrane protein (Putative conserved transmembrane protein) | MSMEG_0441 Uncharacterized protein |
| MSMEG_4324 Transcriptional regulator, CdaR (Uncharacterized protein) | MSMEG_0444 Agmatine deiminase (EC 3.5.3.12) (Putative agmatine deiminase) (EC 3.5.3.12) |
| MSMEG_0646 Phosphate-import permease protein PhnE | MSMEG_2111 Secreted chorismate mutase (CM) (EC 5.4.99.5) |
| MSMEG_1892 Methyltransferase small (Transferase) | MSMEG_0752 Fructose-bisphosphate aldolase (Fructose-bisphosphate aldolase, class II) (EC 4.1.2.13) |
| MSMEG_5050 Methionine aminopeptidase (MAP) (MetAP) (EC 3.4.11.18) (Peptidase M) | MSMEG_0750 Conserved transmembrane protein (Membrane protein) |
| MSMEG_2500 Uncharacterized protein | MSMEG_2353 NmrA-like protein (Secreted protein) |
| MSMEG_3326 Putative glutamate--cysteine ligase 2-2 (EC 6.3.2.2) (Gamma-glutamylcysteine synthetase 2-2) (GCS 2-2) (Gamma-GCS 2-2) | MSMEG_6221 Integral membrane protein (Putative conserved transmembrane protein) |
| MSMEG_3142 HTH-type transcriptional repressor AcnR (Transcriptional regulator, TetR family) | MSMEG_2422 Acylphosphatase (EC 3.6.1.7) (Acylphosphate phosphohydrolase) |
| MSMEG_3144 Putative membrane protein (Uncharacterized protein) | MSMEG_4337 Beta-lactamase |
| MSMEG_0975 ATPases involved in chromosome partitioning (Uncharacterized protein) | MSMEG_4213 Cytochrome P450 hydroxylase |
| MSMEG_3533 Di-/tripeptide transporter | MSMEG_2620 Pterin-4-alpha-carbinolamine dehydratase (EC 4.2.1.96) (Uncharacterized protein) |
| MSMEG_3143 Aconitate hydratase A (ACN) (Aconitase) (EC 4.2.1.3) ((2R,3S)-2-methylisocitrate dehydratase) ((2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate dehydratase) (Iron-responsive protein-like) (IRP-like) (Probable 2-methyl-cis-aconitate hydratase) (EC 4.2.1.99) (RNA-binding protein) | MSMEG_4215 Erythromycin esterase (Phosphoribosyltransferase) |
| MSMEG_6745 Transcriptional regulator, GntR family (Transcriptional regulator, GntR family protein) |
KO_stationary.phase_rep1
WT.vs..DamtR.without.N_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.4.of.6..negative.control
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
X2XR.rep1
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
Wild.type.rep1.x
Biological.replicate.2.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.2.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep1
rel.knockout.rep2
Biological.replicate.6.of.7..panaxydol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
stphcy5plogcy3slide487norm
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.1.of.7..panaxydol.treatment
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
dcpA.knockout.rep1
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X3dbfcy5plogcy3slide601norm
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.2.of.6..isoniazid.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.vs..DglnR.N_starvation_rep2
Wild.type.rep2.x
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X2XR.rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments