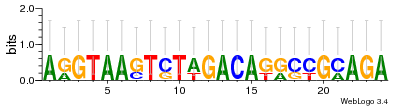
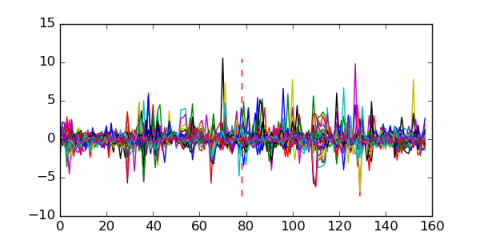
Module 115 Residual: 0.58
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.58 | 0.00000034 | 85 | 78 | 33 | 20160204 |
| MSMEG_6672 Type I phosphodiesterase / nucleotide pyrophosphatase superfamily protein (Type I phosphodiesterase/nucleotide pyrophosphatase) | MSMEG_3031 Shikimate kinase (SK) (EC 2.7.1.71) |
| MSMEG_6671 ABC transporter (Sulfate/thiosulfate import ATP-binding protein CysA) (EC 3.6.3.25) | MSMEG_3739 Cytidylate kinase (CK) (EC 2.7.4.25) (Cytidine monophosphate kinase) |
| MSMEG_6670 ABC transporter, permease protein, putative (Molybdate ABC transporter, permease protein) | MSMEG_3738 GTPase Der (GTP-binding protein EngA) |
| MSMEG_3403 Formamidase (EC 3.5.1.49) (Formamide amidohydrolase) | MSMEG_4699 NAD-specific glutamate dehydrogenase (NAD-GDH) (EC 1.4.1.2) (NAD(+)-dependent glutamate dehydrogenase) |
| MSMEG_3402 Cytosine permease, putative (Permease for cytosine/purines uracil thiamine allantoin) | MSMEG_5351 Uncharacterized protein |
| MSMEG_3401 LamB/YcsF family protein | MSMEG_5583 HNH endonuclease |
| MSMEG_3400 Glutamyl-tRNA(Gln) amidotransferase subunit A (EC 6.3.5.-) | MSMEG_6669 ABC transporter, permease protein |
| MSMEG_6466 Aquaporin Z | MSMEG_0679 Uncharacterized protein |
| MSMEG_3033 3-dehydroquinate synthase (EC 4.2.3.4) | MSMEG_3740 Pseudouridine synthase (EC 5.4.99.-) |
| MSMEG_3030 Chorismate synthase (CS) (EC 4.2.3.5) (5-enolpyruvylshikimate-3-phosphate phospholyase) | MSMEG_3741 Segregation and condensation protein B |
| MSMEG_4179 Conserved integral membrane protein (Probable conserved integral membrane protein) | MSMEG_1151 DNA-binding protein (Transcriptional regulator, XRE family with cupin sensor) |
| MSMEG_4934 ATP:cob(I)alamin adenosyltransferase (EC 2.5.1.17) (EC 4.2.1.28) | MSMEG_3743 SpoOJ regulator protein |
| MSMEG_4932 UDP-N-acetylglucosamine 1-carboxyvinyltransferase (EC 2.5.1.7) (Enoylpyruvate transferase) (UDP-N-acetylglucosamine enolpyruvyl transferase) | MSMEG_0002 6-phosphogluconate dehydrogenase (EC 1.1.1.44) (6-phosphogluconate dehydrogenase, decarboxylating) |
| MSMEG_4668 Oxidoreductase alpha (Molybdopterin) subunit | MSMEG_0003 DNA replication and repair protein RecF |
| MSMEG_3742 Segregation and condensation protein A | MSMEG_0001 DNA polymerase III subunit beta (EC 2.7.7.7) |
| MSMEG_1222 ISMsm6, transposase (Transposase IS4 family protein) | MSMEG_0004 UPF0232 protein MSMEG_0004/MSMEI_0006 |
| MSMEG_3652 CDP-diacylglycerol-glycerol-3-phosphate |
KO_stationary.phase_rep1
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.15min
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
rel.knockout.rep2
Biological.replicate.2.of.7..falcarinol.treatment
dosR.WT_Replicate2
dosR.WT_Replicate1
KO_log.phase_rep2
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Plogcy5plogcy3slide110norm
DglnR_without.vs..with.N_rep1
Biological.replicate.6.of.6..negative.control
MB100pruR.vs..MB100.rep.1
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DglnR.N_starvation_dye.swap_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.1.of.6..kanamycin.treatment
stphcy3plogcy5slide581norm
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
dcpA.knockout.rep1
WT.vs..DamtR.N.surplus_rep1
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.2
Biological.replicate.2.of.6..kanamycin.treatment
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X4XR.rep1
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
X4dbfcy5plogcy3slide901norm
WT.vs..DglnR.N_starvation_rep1
Biological.replicate.1.of.7..falcarinol.treatment
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
Biological.replicate.1.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Plogcy5plogcy3slide113norm
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments