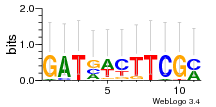
Module 126 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 0.0000000000000014 | 0.000000069 | 74 | 28 | 20160204 |
| MSMEG_1958 PDZ domain family protein | MSMEG_4300 Transcription regulator AmtR (Transcriptional regulator, TetR family) |
| MSMEG_4607 Uncharacterized protein | MSMEG_4302 Adenylate cyclase, putative (CHAD domain containing protein) |
| MSMEG_3526 Uncharacterized protein | MSMEG_3674 Uncharacterized protein |
| MSMEG_0536 Intracellular protease, PfpI family (Intracellular protease, PfpI family protein) | MSMEG_2820 Uncharacterized protein |
| MSMEG_0725 IS1137, transposase orfB | MSMEG_5475 Acetate operon repressor (IclR family transcriptional regulator) |
| MSMEG_0723 Uncharacterized protein | MSMEG_4597 Putative conserved lipoprotein LppH (EC 2.7.11.1) (Putative conserved lipoprotein lpph) |
| MSMEG_0452 Gluconate permease (Inner membrane permease YgbN) | MSMEG_0630 Trehalose 2-sulfotransferase (EC 2.8.2.37) |
| MSMEG_1248 Uncharacterized protein | MSMEG_6227 Transcriptional regulator, PadR family protein |
| MSMEG_3944 LuxR family two-component response regulator (Two component transcriptional regulatory protein devr) | MSMEG_6040 Uncharacterized protein |
| MSMEG_3807 TetR-type regulator | MSMEG_3818 Glutaminase (EC 3.5.1.2) |
| MSMEG_3943 Uncharacterized protein | MSMEG_2280 Pyruvate dehydrogenase (Thiamine pyrophosphate-requiring enzyme) |
| MSMEG_3821 Amino acid permease | MSMEG_2914 L-idonate 5-dehydrogenase (EC 1.1.1.264) |
| MSMEG_3942 Uncharacterized protein | MSMEG_0276 Aldehyde-alcohol dehydrogenase (EC 1.1.1.1) (EC 1.2.1.10) |
| MSMEG_4301 Acyl-CoA synthase | MSMEG_2912 Inner membrane metabolite transport protein YdfJ |
KO_stationary.phase_rep2
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep2
Biological.replicate.4.of.6..negative.control
X4XR.rep1
Bedaquiline.vs..DMSO.30min
Biological.replicate.4.of.6..isoniazid.treatment
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate2
Biological.replicate.5.of.6..negative.control
KO_log.phase_rep2
KO_log.phase_rep1
Wild.type.rep1.y
Biological.replicate.6.of.6..negative.control
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.5.of.7..panaxydol.treatment
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.3.of.7..falcarinol.treatment
Wild.type.rep3
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
WT_log.phase_rep2
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
Biological.replicate.5.of.6..kanamycin.treatment
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.6.of.7..ethambutol.treatment
WT.vs..DamtR.N.surplus_rep1
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy5plogcy3slide487norm
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.1.of.7..panaxydol.treatment
dcpA.knockout.rep2
Biological.replicate.2.of.6..isoniazid.treatment
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.4.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.6.of.6..kanamycin.treatment
Bedaquiline.vs..DMSO.60min
Biological.replicate.2.of.6..kanamycin.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
Biological.replicate.3.of.6..kanamycin.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
X2XR.rep1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.vs..DglnR.N_starvation_rep1
Plogcy5plogcy3slide113norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments