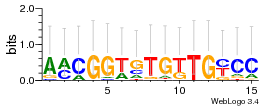
Module 136 Residual: 0.51
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.51 | 9e-22 | 0.0000022 | 77 | 34 | 20160204 |
| MSMEG_6482 Secreted protein | MSMEG_5665 Uncharacterized protein |
| MSMEG_1886 Fatty acid desaturase (EC 1.14.19.3) | MSMEG_4006 Transcriptional regulator, CdaR, putative |
| MSMEG_5082 DNA-3-methyladenine glycosylase (DNA-3-methyladenine glycosylase I) (EC 3.2.2.20) | MSMEG_4007 Oxidoreductase, 2OG-Fe(II) oxygenase family protein |
| MSMEG_3318 Monooxygenase FAD-binding protein (Oxidoreductase) | MSMEG_5230 Uncharacterized protein |
| MSMEG_5085 Dihydropteroate synthase (DHPS) (EC 2.5.1.15) (Dihydropteroate pyrophosphorylase) | MSMEG_5988 Uncharacterized protein |
| MSMEG_6504 Uncharacterized protein | MSMEG_5989 PPE family protein (Uncharacterized protein) |
| MSMEG_6771 Uncharacterized protein | MSMEG_6850 Alpha/beta hydrolase (Alpha/beta hydrolase fold protein) (EC 3.3.2.9) |
| MSMEG_6502 Uncharacterized protein | MSMEG_0712 Transcriptional regulator, TetR family (Transcriptional regulator, TetR family protein) |
| MSMEG_6792 Gluconate permease (Inner membrane permease YgbN) | MSMEG_1563 Short-chain dehydrogenase/reductase SDR |
| MSMEG_5346 TetR-family protein transcriptional regulator | MSMEG_5372 Sensor protein KdpD (EC 2.7.13.3) |
| MSMEG_5347 Major facilitator superfamily MFS_1 (Transporter, major facilitator family protein) | MSMEG_6206 Serine/threonine protein kinase PksC (EC 2.7.11.1) (Serine/threonine-protein kinase) |
| MSMEG_6479 Putative transcriptional regulator | MSMEG_6207 Uncharacterized protein |
| MSMEG_4240 Polyprenyl synthetase | MSMEG_5420 Tat-translocated enzyme |
| MSMEG_4241 Possible conserved integral membrane protein (Putative conserved integral membrane protein) | MSMEG_6585 Acyl-CoA dehydrogenase |
| MSMEG_2592 Conserved hypothetical membrane protein (Uncharacterized protein) | MSMEG_6584 Acyl-CoA dehydrogenase |
| MSMEG_2591 Uncharacterized protein | MSMEG_4096 Nitrilotriacetate monooxygenase (Nitrilotriacetate monooxygenase component A) (EC 1.14.13.-) |
| MSMEG_4611 Carrier protein (Dicarboxylate carrier MatC domain protein) | MSMEG_6582 FAD-dependent pyridine nucleotide-disulfide oxidoreductase (Pyridine nucleotide-disulphide oxidoreductase domain protein) |
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.3.of.6..kanamycin.treatment
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Plogcy5plogcy3slide110norm
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.1.of.6..kanamycin.treatment
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
rel.knockout.rep2
Wild.type.rep3
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.3.of.6..isoniazid.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
Sub.MIC.INH
X0.6..vs..50..air.saturation.Replicate.4
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.3
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.2.of.7..ethambutol.treatment
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
X3dbfcy5plogcy3slide601norm
stphcy3plogcy5slide581norm
DglnR_without.vs..with.N_rep1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X2.5..vs..50..air.saturation.Replicate.2
WT_stationary.phase_rep2
WT_stationary.phase_rep1
X4XR.rep1
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_dyeswap_rep2
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
X3dbfcy5plogcy3slide603norm
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments