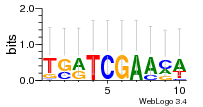
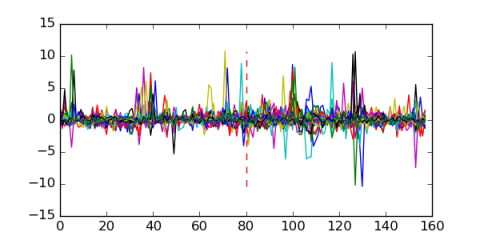
Module 139 Residual: 0.51
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.51 | 0.00000000000041 | 0.000017 | 80 | 30 | 20160204 |
| MSMEG_4422 Oxidoreductase | MSMEG_2985 Fumarate hydratase class (EC 4.2.1.2) (Fumarate hydratase class I, anaerobic) |
| MSMEG_4423 Alcohol dehydrogenase, zinc-binding protein (EC 1.6.5.5) (Oxidoreductase) | MSMEG_6760 Activator of Hsp90 ATPase 1-like protein (Uncharacterized protein) |
| MSMEG_4730 Uncharacterized protein | MSMEG_5278 Conserved membrane-spanning protein |
| MSMEG_5975 Methyltransferase type 12, putative (Uncharacterized protein) | MSMEG_6762 Transcriptional regulator |
| MSMEG_4728 Acyltransferase papA2 (Condensation domain protein) | MSMEG_2343 Methylesterase |
| MSMEG_1387 Acyl-CoA dehydrogenase (Putative acyl-CoA dehydrogenase) | MSMEG_4729 Uncharacterized protein |
| MSMEG_1388 Enoyl-CoA hydratase (Enoyl-CoA hydratase EchA4) (EC 4.2.1.17) | MSMEG_3798 Uncharacterized protein |
| MSMEG_1389 Uncharacterized protein | MSMEG_2347 Phytoene dehydrogenase (EC 1.14.99.-) (Phytoene desaturase) (EC 5.4.99.9) |
| MSMEG_2633 ABC transporter ATP-binding protein (ABC-type transporter, ATP-binding protein) (EC 3.6.3.36) | MSMEG_2346 Phytoene synthase (EC 2.5.1.-) (EC 2.5.1.32) |
| MSMEG_2841 Putative monooxygenase | MSMEG_2344 Dehydrogenase |
| MSMEG_2635 ABC transporter permease protein (Binding-protein-dependent transport systems inner membrane component) | MSMEG_1390 Enoyl-CoA hydratase (Enoyl-CoA hydratase/isomerase) (EC 4.2.1.17) |
| MSMEG_2634 Uncharacterized protein | MSMEG_4193 Phosphoglycerate mutase (EC 5.4.2.-) (Phosphoglycerate mutase family protein) |
| MSMEG_0271 Uncharacterized protein | MSMEG_4194 Undecaprenyl-diphosphatase (EC 3.6.1.27) (Bacitracin resistance protein) (Undecaprenyl pyrophosphate phosphatase) |
| MSMEG_0273 Ethanolamine utilization protein EutN (Ethanolamine utilization protein EutN/carboxysome structural protein CcmL) | MSMEG_3480 Aminoglycoside/hydroxyurea antibiotic resistance kinase (Streptomycin 6-kinase (Streptidine kinase) (Streptomycin6-phosphotransferase) (APH(6))) (EC 2.7.1.72) |
| MSMEG_0272 Microcompartments protein (Propanediol utilization protein PduA) | MSMEG_3479 Probable thiol peroxidase (EC 1.11.1.-) |
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
X4XR.rep1
dcpA.knockout.rep2
Biological.replicate.4.of.6..negative.control
Wild.type.rep2.x
Biological.replicate.5.of.6..isoniazid.treatment
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate1
KO_log.phase_rep2
DglnR_without.vs..with.N_rep1
Biological.replicate.7.of.7..falcarinol.treatment
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
Sub.MIC.INH
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.2.of.7..falcarinol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
stphcy5plogcy3slide487norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
Plogcy5plogcy3slide111norm
Biological.replicate.2.of.6..isoniazid.treatment
X3dbfcy5plogcy3slide601norm
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy3plogcy5slide581norm
WT_log.phase_rep1
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
dosR.WT_Replicate3
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Biological.replicate.3.of.6..kanamycin.treatment
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.N.surplus_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments