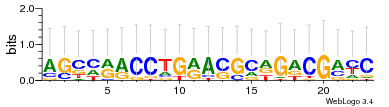
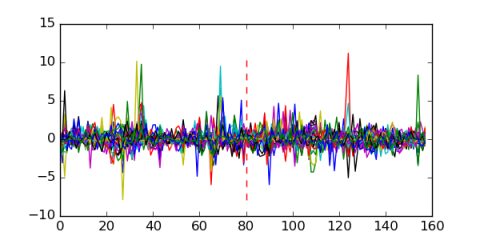
Module 142 Residual: 0.65
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.65 | 4.8e-16 | 0.00000013 | 80 | 30 | 20160204 |
| MSMEG_4352 Putative secreted protein | MSMEG_2772 Amino acid permease (Putative conserved integral membrane alanine and valine and leucine rich protein (Amino acid permease, putative)) |
| MSMEG_4353 Uncharacterized protein | MSMEG_2773 Conserved alanine valine and glycine rich protein (Putative RNA methyltransferase) (EC 2.1.1.-) |
| MSMEG_4356 Binding-protein-dependent transport systems inner membrane component (Inner membrane ABC transporter permease protein YddQ) | MSMEG_2771 TrkA protein (TrkA-N) |
| MSMEG_4357 ABC transporter related protein (ABC transporter, ATP-binding protein) | MSMEG_2658 Beta-lactamase (EC 3.5.2.6) |
| MSMEG_4354 Dipeptide-binding protein of ABC transport system | MSMEG_3732 Uncharacterized protein |
| MSMEG_4355 Peptide ABC transporter, permease protein | MSMEG_3068 Uncharacterized protein |
| MSMEG_4358 D-beta-hydroxybutyrate dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) | MSMEG_3063 LemA protein |
| MSMEG_5615 Uncharacterized protein | MSMEG_3004 Hemerythrin HHE cation binding domain protein (Uncharacterized protein) |
| MSMEG_1722 Uncharacterized protein | MSMEG_5725 PLP-dependent aminotransferases (Succinyldiaminopimelate aminotransferase) (EC 2.6.1.11) |
| MSMEG_3686 Uncharacterized protein | MSMEG_6661 O-methyltransferase Omt (O-methyltransferase, putative) |
| MSMEG_5613 Probable conserved transmembrane protein | MSMEG_1077 Uncharacterized protein |
| MSMEG_2416 Uncharacterized protein | MSMEG_1566 Oxidoreductase |
| MSMEG_0885 Twin-arginine translocation pathway signal (Uncharacterized protein) | MSMEG_3180 Regulatory protein, MerR (Transcriptional regulator, MerR family protein) |
| MSMEG_2657 Peptidase, M16 family protein | MSMEG_1723 Uncharacterized protein |
| MSMEG_2656 Polyribonucleotide nucleotidyltransferase (EC 2.7.7.8) (Polynucleotide phosphorylase) (PNPase) | MSMEG_2769 TrkA-N (TrkB protein) |
Wild.type.rep2.x
KO_stationary.phase_rep1
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Biological.replicate.5.of.7..panaxydol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Wild.type.rep3
MB100pruR.vs..MB100..rep.3
Biological.replicate.5.of.6..negative.control
KO_log.phase_rep2
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
WT_log.phase_rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DglnR.N_starvation_dye.swap_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.3
X3dbfcy3plogcy5slide607norm
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
stphcy3plogcy5slide581norm
WT_log.phase_rep1
X3dbfcy5plogcy3slide603norm
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..negative.control
WT_stationary.phase_rep1
Biological.replicate.2.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
dosR.WT_Replicate3
X4dbfcy5plogcy3slide480norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.N.surplus_dyeswap_rep2
DglnR_without.vs..with.N_dyeswap_rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DglnR.N_starvation_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..falcarinol.treatment
Comments
Log in to post comments