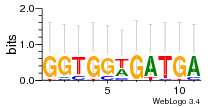
Module 179 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 0.000000000078 | 0.0000000065 | 82 | 29 | 20160204 |
| MSMEG_3176 RarD protein | MSMEG_3196 Uncharacterized protein |
| MSMEG_3175 Pseudouridine synthase (EC 5.4.99.-) | MSMEG_3290 Regulatory protein (Regulatory protein LuxR) |
| MSMEG_3174 Lipoprotein signal peptidase (EC 3.4.23.36) (Prolipoprotein signal peptidase) (Signal peptidase II) | MSMEG_4385 ABC transporter oligopeptide binding protein |
| MSMEG_1880 Hypothetical alanine arginine proline rich protein | MSMEG_4387 ABC transporter permease protein (Binding-protein-dependent transport systems inner membrane component) |
| MSMEG_1882 Diacylglycerol O-acyltransferase (EC 2.3.1.20) | MSMEG_4386 ABC transporter permease protein (Oligopeptide ABC transporter, permease) (EC 3.6.3.24) |
| MSMEG_5712 LysR-family protein transcriptional regulator (LysR-family transcriptional regulator) | MSMEG_4674 Trigger factor (TF) (EC 5.2.1.8) (PPIase) |
| MSMEG_5714 Short-chain dehydrogenase/reductase SDR (EC 1.1.1.100) | MSMEG_1638 Roadblock/LC7 domain protein (Roadblock/LC7 family protein) |
| MSMEG_4391 Acyl-CoA dehydrogenase (EC 1.3.8.-) (Acyl-CoA dehydrogenase-family protein) | MSMEG_1639 Uncharacterized protein |
| MSMEG_0415 Flavin reductase-like, FMN-binding protein (NADH-fmn oxidoreductase) | MSMEG_1637 Histidine kinase (EC 2.7.13.3) |
| MSMEG_0414 Oxidoreductase, 2OG-Fe(II) oxygenase family protein | MSMEG_4389 Coenzyme F420-dependent N5,N10-methylene tetrahydromethanopterin reductase-like flavin-dependent oxidoreductase (EC 1.14.14.5) (Monooxygenase, NtaA/SnaA/SoxA family protein) |
| MSMEG_1641 Pentapeptide repeat protein MfpA (Mycobacterial fluoroquinone resistance pentapeptide) (Orf3) | MSMEG_4388 ABC transporter (ABC transporter ATP-binding protein) |
| MSMEG_1640 ATP/GTP binding protein (ATP/GTP-binding protein) | MSMEG_0614 Putative S-adenosyl-L-methionine-dependent methyltransferase MSMEG_0614/MSMEI_0598 (EC 2.1.1.-) |
| MSMEG_1688 Cupin 2, conserved barrel domain protein (Cupin domain protein) | MSMEG_2509 Mg-chelatase subunits D/I family protein, ComM subfamily protein |
| MSMEG_0607 Methyltransferase, putative, family protein | MSMEG_4913 LpqM protein |
| MSMEG_3195 Uncharacterized protein |
Wild.type.rep1.y
KO_stationary.phase_rep1
WT.vs..DglnR.N_starvation_rep2
KO_log.phase_rep1
X4XR.rep1
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.3.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
Wild.type.rep2.y
Plogcy5plogcy3slide110norm
X3dbfcy5plogcy3slide601norm
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
DglnR_without.vs..with.N_rep2
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
Wild.type.rep3
Bedaquiline.vs..DMSO..45min
M..smegmatis.Acid.NO.treated
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.4
WT.vs..DamtR.N.surplus_rep1
stphcy5plogcy3slide487norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.4.of.6..negative.control
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
X4XR.rep2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
Biological.replicate.3.of.6..kanamycin.treatment
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.with.vs..without.N_rep2
Wild.type.rep1.x
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
DglnR_without.vs..with.N_dyeswap_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
WT.vs..DglnR.N_starvation_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Sub.MIC.INH
Biological.replicate.2.of.6..negative.control
BatchCulture_LBTbroth_StationaryPhase_Replicate1
dosR.WT_Replicate2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments