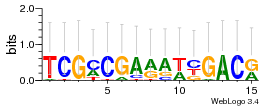
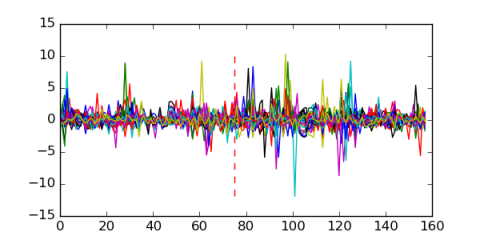
Module 182 Residual: 0.54
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.54 | 0.00000000000014 | 0.000000000000036 | 75 | 34 | 20160204 |
| MSMEG_3726 Alcohol dehydrogenase (PQQ-dependent dehydrogenase, methanol/ethanol family) | MSMEG_5298 Uncharacterized protein |
| MSMEG_4540 Acetyltransferase, gnat family protein (GCN5-related N-acetyltransferase) | MSMEG_0877 Dihydrodipicolinate synthase (EC 4.3.3.7) |
| MSMEG_2209 Transcriptional regulator, GntR family (Transcriptional regulator, GntR family protein) | MSMEG_3130 Uncharacterized protein |
| MSMEG_4070 Transcriptional regulator, TetR family protein, putative (Transcriptional regulatory protein) | MSMEG_5454 Choloylglycine hydrolase (EC 3.5.1.24) (Choloylglycine hydrolase, putative) |
| MSMEG_4406 UsfY protein | MSMEG_5980 Methyltransferase (NDP-hexose 3-C-methyltransferase protein) |
| MSMEG_2042 Aminoglycoside phosphotransferase (Phosphotransferase enzyme family protein) | MSMEG_5453 Uncharacterized protein |
| MSMEG_2043 Transcriptional regulator, TetR family protein (Transcriptional regulatory protein (Probably TetR-family)) | MSMEG_2208 Acyl-CoA dehydrogenase (EC 1.3.8.1) (EC 1.3.99.-) |
| MSMEG_4408 Integral membrane protein (Putative membrane spanning protein) | MSMEG_5565 Uncharacterized protein |
| MSMEG_0531 Acyl-CoA dehydrogenase (EC 1.3.8.-) (Putative acyl-CoA dehydrogenase) | MSMEG_2206 3-oxoacyl-(Acyl-carrier-protein) reductase (EC 1.1.1.100) |
| MSMEG_2534 Putative carboxylesterase protein | MSMEG_2207 Beta-ketothiolase |
| MSMEG_2535 Dehydrogenase/reductase SDR family protein member 10 (EC 1.1.-.-) | MSMEG_4539 Alkanesulfonate monooxygenase (EC 1.14.14.5) (Alkanesulfonate monooxygenase 2) |
| MSMEG_2536 3-oxoacyl-[acyl-carrier-protein] reductase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) | MSMEG_2241 AMP-dependent synthetase and ligase (Medium chain acyl-CoA synthetase) |
| MSMEG_5976 Epimerase/dehydratase (NAD dependent epimerase/dehydratase) (EC 5.1.3.2) | MSMEG_3181 Uncharacterized protein |
| MSMEG_2533 Uncharacterized protein | MSMEG_3484 Cupin domain protein (Uncharacterized protein) |
| MSMEG_5979 Putative glucose-1-phosphate cytidylyltransferase (EC 2.7.7.33) (Transferase) | MSMEG_2205 Putative acyl-CoA dehydrogenase |
| MSMEG_5978 GlcNAc-PI de-N-acetylase family protein (Uncharacterized protein) | MSMEG_0514 Alpha-galactosidase (EC 3.2.1.22) |
| MSMEG_4409 Uncharacterized protein | MSMEG_4067 Zinc-containing alcohol dehydrogenase superfamily (EC 1.6.5.5) (Zinc-containing alcohol dehydrogenase superfamily protein) |
KO_stationary.phase_rep2
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
Wild.type.rep2.x
Bedaquiline.vs..DMSO.30min
Biological.replicate.1.of.6..negative.control
Wild.type.rep3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.2.of.6..kanamycin.treatment
KO_log.phase_rep1
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.y
Biological.replicate.3.of.7..panaxydol.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Biological.replicate.6.of.7..panaxydol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
X0.6..vs..50..air.saturation.Replicate.1
Biological.replicate.5.of.7..falcarinol.treatment
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.2.of.7..falcarinol.treatment
mc2155.vs.Dcrp1.replicate.2
Plogcy5plogcy3slide113norm
Biological.replicate.7.of.7..panaxydol.treatment
stphcy3plogcy5slide581norm
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
X4dbfcy5plogcy3slide480norm
Biological.replicate.2.of.6..isoniazid.treatment
WT_log.phase_rep2
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
X2.5..vs..50..air.saturation.Replicate.1
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
mc2155.vs.Dcrp1.replicate.1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.with.vs..without.N_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_rep1
X2XR.rep1
Biological.replicate.1.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
BatchCulture_LBTbroth_StationaryPhase_Replicate2
dosR.WT_Replicate2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments