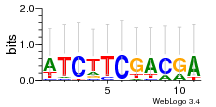
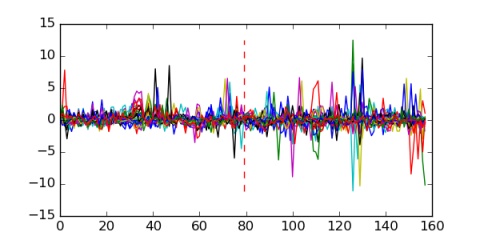
Module 195 Residual: 0.53
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.53 | 0.000000001 | 2.9 | 79 | 31 | 20160204 |
| MSMEG_6673 6-aminohexanoate-cyclic-dimer hydrolase (EC 3.5.2.12) (Amidase) (EC 3.5.1.4) | MSMEG_6705 IclR family transcriptional regulator (Regulatory protein) |
| MSMEG_6885 Coenzyme F420-dependent N5 N10-methylene tetrahydromethanopterin reductase-like protein (MmcI protein) | MSMEG_4109 NAD(P) transhydrogenase, alpha subunit (EC 1.6.1.1) |
| MSMEG_0680 UDP-glucose 6-dehydrogenase (EC 1.1.1.22) | MSMEG_4346 Uncharacterized protein |
| MSMEG_1840 Rubredoxin | MSMEG_4453 Uncharacterized protein |
| MSMEG_1842 Transcriptional regulator, TetR family protein | MSMEG_6640 Uncharacterized protein |
| MSMEG_0455 Aldehyde Dehydrogenase (EC 1.2.1.-) (Aldehyde dehydrogenase) | MSMEG_5720 Enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35) (Putative 3-hydroxyacyl-CoA dehydrogenase) |
| MSMEG_4951 50S ribosomal protein L31 | MSMEG_5376 Uncharacterized protein |
| MSMEG_1668 Pyridoxamine 5'-phosphate oxidase family protein | MSMEG_2521 Amidase (Putative amidase amiC) (EC 3.5.1.4) |
| MSMEG_4108 NAD(P) transhydrogenase subunit beta (EC 1.6.1.2) (Nicotinamide nucleotide transhydrogenase subunit beta) | MSMEG_6571 Uncharacterized protein |
| MSMEG_2025 Putative IclR family protein transcriptional regulator (Transcriptional regulator IclR) | MSMEG_3483 Mosc domain protein (Putative MOSC domain protein) |
| MSMEG_5118 NUDIX hydrolase (EC 3.6.1.13) (Nudix hydrolase) | MSMEG_2965 GTP pyrophosphokinase (EC 2.7.6.5) (GTP pyrophosphokinase RelA) (EC 2.7.6.5) |
| MSMEG_2714 Uncharacterized protein | MSMEG_3481 o-succinylbenzoate synthase (EC 4.2.1.113) |
| MSMEG_2710 Uncharacterized protein | MSMEG_1841 Rubredoxin |
| MSMEG_2711 (NiFe) hydrogenase maturation protein HypF ([NiFe] hydrogenase maturation protein HypF) | MSMEG_2708 Uncharacterized protein |
| MSMEG_2712 Hydrogenase assembly chaperone HypC/HupF (Hydrogenase assembly chaperone hypC/hupF) | MSMEG_1839 Alkane 1-monooxygenase (EC 1.14.15.3) (Putative transmembrane alkane 1-monooxygenase Alkb) (EC 1.14.15.3) |
| MSMEG_2713 Peptidase M52, hydrogen uptake protein (Putative NiFe hydrogenase-specific C-terminal protease) |
Biological.replicate.5.of.7..falcarinol.treatment
KO_log.phase_rep1
Biological.replicate.4.of.6..negative.control
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.3.of.7..ethambutol.treatment
MB100pruR.vs..MB100..rep.3
KO_log.phase_rep2
Wild.type.rep2.y
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.y
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.3
WT.vs..DglnR.N_starvation_dye.swap_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
stphcy5plogcy3slide487norm
Biological.replicate.1.of.6..kanamycin.treatment
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
Plogcy5plogcy3slide111norm
X3dbfcy5plogcy3slide601norm
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
WT_log.phase_rep1
DglnR_without.vs..with.N_rep1
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.4.of.6..isoniazid.treatment
mc2155.vs.Dcrp1.replicate.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
Biological.replicate.5.of.6..kanamycin.treatment
Wild.type.rep2.x
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.3.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments