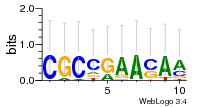
Module 214 Residual: 0.60
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.60 | 0.000003 | 0.0036 | 78 | 35 | 20160204 |
| MSMEG_5218 Putative ester cyclase (Uncharacterized protein) | MSMEG_5193 Transcriptional regulator, AraC family protein |
| MSMEG_5678 Glyoxalase/bleomycin resistance protein/dioxygenase (EC 4.4.1.5) | MSMEG_1736 Glycerol-3-phosphate dehydrogenase (EC 1.1.5.3) |
| MSMEG_5679 Putative DNA-binding protein (Transcriptional regulator, ArsR family) | MSMEG_2997 Acyl-CoA thioesterase (Uncharacterized protein) |
| MSMEG_6930 Uncharacterized protein | MSMEG_3576 Alpha amylase catalytic region (EC 3.2.1.54) (Alpha-amylase 3) |
| MSMEG_4548 Acetyltransferase, GNAT family (Acetyltransferase, gnat family protein) | MSMEG_3575 CMP/dCMP deaminase, zinc-binding protein (Cytidine/deoxycytidylate deaminase, zinc-binding region) |
| MSMEG_4662 Deoxyribose-phosphate aldolase superfamily protein | MSMEG_6768 Halogenase |
| MSMEG_4663 Myo-inositol catabolism IolB protein (Protein IolB) | MSMEG_0321 Transcriptional regulator (Uncharacterized protein) |
| MSMEG_5304 Two-component sensor kinase | MSMEG_6520 Orotate phosphoribosyltransferase (OPRT) (OPRTase) (EC 2.4.2.10) |
| MSMEG_4666 Inositol 2-dehydrogenase (EC 1.1.1.18) (Myo-inositol 2-dehydrogenase) (MI 2-dehydrogenase) | MSMEG_3559 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) |
| MSMEG_4664 IolD protein (Thiamine pyrophosphate-requiring enzyme, putative acetolactate synthase) (EC 2.2.1.6) | MSMEG_5680 Glyoxalase family protein (Glyoxalase/bleomycin resistance protein/dioxygenase) |
| MSMEG_4665 IolE protein (Xylose isomerase domain protein TIM barrel) | MSMEG_1401 Elongation factor Tu (EF-Tu) |
| MSMEG_0600 Dehydrogenase (Short-chain dehydrogenase/reductase SDR) | MSMEG_3892 Uncharacterized protein |
| MSMEG_6858 Epoxide hydrolase (Epoxide hydrolase 1) (EC 3.3.2.9) | MSMEG_3893 Uncharacterized protein |
| MSMEG_0170 Major facilitator superfamily MFS_1 (Transmembrane transport protein) | MSMEG_1446 NTP pyrophosphohydrolase |
| MSMEG_4320 Alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal allergen (EC 1.11.1.15) | MSMEG_5306 Transcriptional regulatory protein |
| MSMEG_4617 Glutamine-dependent NAD(+) synthetase (EC 6.3.5.1) (Glutamine-dependent NAD+ synthetase) (EC 6.3.5.1) | MSMEG_0169 Uncharacterized protein |
| MSMEG_4321 Uncharacterized protein | MSMEG_5289 Uncharacterized protein |
| MSMEG_3354 TetR family protein transcriptional regulatory protein (TetR-family transcriptional regulator) |
KO_stationary.phase_rep1
WT.vs..DamtR.without.N_rep1
Biological.replicate.2.of.7..panaxydol.treatment
X4XR.rep1
Bedaquiline.vs..DMSO.15min
Mycobacterium.smegmatis.Spheroplast.induced
Wild.type.rep2.x
Biological.replicate.3.of.7..falcarinol.treatment
Wild.type.rep1.x
Wild.type.rep3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
X3dbfcy5plogcy3slide601norm
Wild.type.rep1.y
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
MB100pruR.vs..MB100.rep.1
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
DamtR.with.vs..DamtR.without.N_.rep2
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.5
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.7.of.7..panaxydol.treatment
X3dbfcy3plogcy5slide607norm
dcpA.knockout.rep2
dcpA.knockout.rep1
Plogcy5plogcy3slide111norm
Biological.replicate.5.of.6..negative.control
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Biological.replicate.5.of.6..isoniazid.treatment
Bedaquiline.vs..DMSO.60min
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
WT.vs..DamtR.N.surplus_dyeswap_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.2.of.6..negative.control
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
Biological.replicate.6.of.7..panaxydol.treatment
dosR.WT_Replicate2
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments