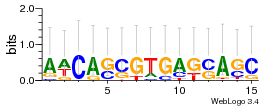
Module 239 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 0.000000000000022 | 0.00000000013 | 78 | 31 | 20160204 |
| MSMEG_1958 PDZ domain family protein | MSMEG_2358 tRNA-specific 2-thiouridylase MnmA (EC 2.8.1.-) |
| MSMEG_0784 Acetate kinase (EC 2.7.2.1) (Acetokinase) | MSMEG_2901 Uncharacterized protein |
| MSMEG_1957 Uncharacterized protein | MSMEG_6080 DNA integrity scanning protein DisA (Cyclic-di-AMP synthase) (c-di-AMP synthase) (Diadenylate cyclase) (EC 2.7.7.85) |
| MSMEG_2180 RemK protein (S-isoprenylcysteine O-methyltransferase-like protein) (EC 2.1.1.100) | MSMEG_3651 Membrane associated protein (Uncharacterized protein) |
| MSMEG_0783 Phosphate acetyltransferase (Putative phosphate acetyltransferase Pta (Phosphotransacetylase)) (EC 2.3.1.8) | MSMEG_0365 Uncharacterized protein |
| MSMEG_2182 TetR family regulator (Transcriptional regulator) | MSMEG_0364 Uncharacterized protein |
| MSMEG_5227 Exodeoxyribonuclease 7 small subunit (EC 3.1.11.6) (Exodeoxyribonuclease VII small subunit) | MSMEG_0508 Glycerol-phosphate porter (Sugar ABC transporter ATP-binding protein) |
| MSMEG_5226 Exodeoxyribonuclease 7 large subunit (EC 3.1.11.6) | MSMEG_6188 Putative membrane protein (Uncharacterized protein) |
| MSMEG_3788 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase (EC 4.1.3.16) (KHG/KDPG aldolase) (EC 4.1.2.14) | MSMEG_3089 Deoxyribose-phosphate aldolase (DERA) (EC 4.1.2.4) (2-deoxy-D-ribose 5-phosphate aldolase) (Phosphodeoxyriboaldolase) |
| MSMEG_3789 Inner membrane permease YgbN (Low-affinity gluconate transporter) | MSMEG_6187 Endonuclease III (EC 4.2.99.18) (DNA-(apurinic or apyrimidinic site) lyase) |
| MSMEG_1559 Phosphoglucosamine mutase (EC 5.4.2.10) | MSMEG_1276 Uncharacterized protein MSMEG_1276/MSMEI_1238.1 |
| MSMEG_3787 D-Aminoacylase (EC 3.5.1.-) (D-aminoacylase) (EC 3.5.1.81) | MSMEG_1275 HNH nuclease, putative |
| MSMEG_2498 Uncharacterized protein | MSMEG_2580 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase (flavodoxin) (EC 1.17.7.3) (1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase) |
| MSMEG_0763 Antibiotic transporter | MSMEG_0267 Esterase |
| MSMEG_2357 Cysteine desulfurase (EC 2.8.1.7) | MSMEG_0265 Uracil DNA glycosylase superfamily protein |
| MSMEG_0764 Na+/H+ antiporter |
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Sub.MIC.INH
Biological.replicate.4.of.6..negative.control
X4XR.rep1
Bedaquiline.vs..DMSO.30min
Biological.replicate.3.of.7..ethambutol.treatment
Wild.type.rep3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.2.of.6..kanamycin.treatment
Wild.type.rep2.y
Wild.type.rep1.y
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.3.of.6..kanamycin.treatment
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.7..panaxydol.treatment
M..smegmatis.Acid.NO.treated
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
mc2155.vs.Dcrp1.replicate.1
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.2.of.7..falcarinol.treatment
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
Biological.replicate.2.of.7..ethambutol.treatment
WT_log.phase_rep1
X4dbfcy5plogcy3slide480norm
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.2.of.6..negative.control
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments