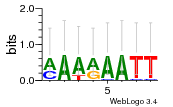
Module 250 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 4.2e-30 | 0.000011 | 80 | 24 | 20160204 |
| MSMEG_5126 FO synthase (EC 2.5.1.-) | MSMEG_0188 MarR-family protein transcriptional regulator |
| MSMEG_5127 Uncharacterized protein | MSMEG_3553 D-hydantoinase (Dihydropyrimidinase) (Dihydropyrimidinase) (EC 3.5.2.2) |
| MSMEG_5322 Uncharacterized protein | MSMEG_2202 Dimethylaniline monooxygenase [N-oxide-forming] 5 (EC 1.14.13.8) (Monooxygenase, flavin-binding protein) (EC 1.14.13.8) |
| MSMEG_1649 FAD dependent oxidoreductase (Monooxygenase FAD-binding protein) (EC 1.14.13.50) | MSMEG_0186 MmpS2 (MmpS2 protein) |
| MSMEG_1648 Putative transcriptional regulator | MSMEG_0400 Peptide synthetase |
| MSMEG_4648 DNA-binding protein (Regulatory protein LacI) | MSMEG_0301 Oxidoreductase, FAD-binding (Putative glycolate oxidase FAD-linked subunit) (EC 1.1.2.4) |
| MSMEG_1646 23S rRNA methyltransferase (Ribosomal RNA adenine dimethylase family protein) | MSMEG_5928 Uncharacterized protein |
| MSMEG_1478 Lysine exporter protein (LYSE/YGGA) (Transporter, LysE family protein) | MSMEG_0200 AAA ATPase containing von Willebrand factor type A (VWA) domain-like protein (Uncharacterized protein) |
| MSMEG_3095 D-ribose-binding periplasmic protein (EC 3.6.3.17) | MSMEG_4191 Uncharacterized protein |
| MSMEG_3097 Phosphoenolpyruvate carboxylase (PEPC) (PEPCase) (EC 4.1.1.31) | MSMEG_4192 Uncharacterized protein |
| MSMEG_3554 Methylenetetrahydromethanopterin reductase (EC 1.5.99.11) (N5,N10-methylene-tetrahydromethanopterin reductase) | MSMEG_5143 HTH-type transcriptional regulator DegA (LacI-family transcriptional regulator) (EC 5.1.1.1) |
| MSMEG_0303 Alcohol dehydrogenase, zinc-containing | MSMEG_5144 Glycoside hydrolase, family 3-like protein (EC 3.2.1.21) (Xylosidase/arabinosidase) |
Bedaquiline.vs..DMSO.30min
WT.vs..DamtR.without.N_rep1
Biological.replicate.2.of.7..panaxydol.treatment
dcpA.knockout.rep2
Biological.replicate.4.of.6..kanamycin.treatment
Biological.replicate.2.of.6..isoniazid.treatment
Biological.replicate.1.of.6..negative.control
Wild.type.rep3
KO_log.phase_rep2
KO_log.phase_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Bedaquiline.vs..DMSO.60min
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep2
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
stphcy5plogcy3slide487norm
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.6.of.7..ethambutol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
X4dbfcy5plogcy3slide901norm
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide601norm
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
X3dbfcy5plogcy3slide603norm
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..negative.control
Biological.replicate.1.of.7..ethambutol.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.2
Biological.replicate.3.of.6..kanamycin.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DglnR.N_starvation_rep2
X2XR.rep1
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments